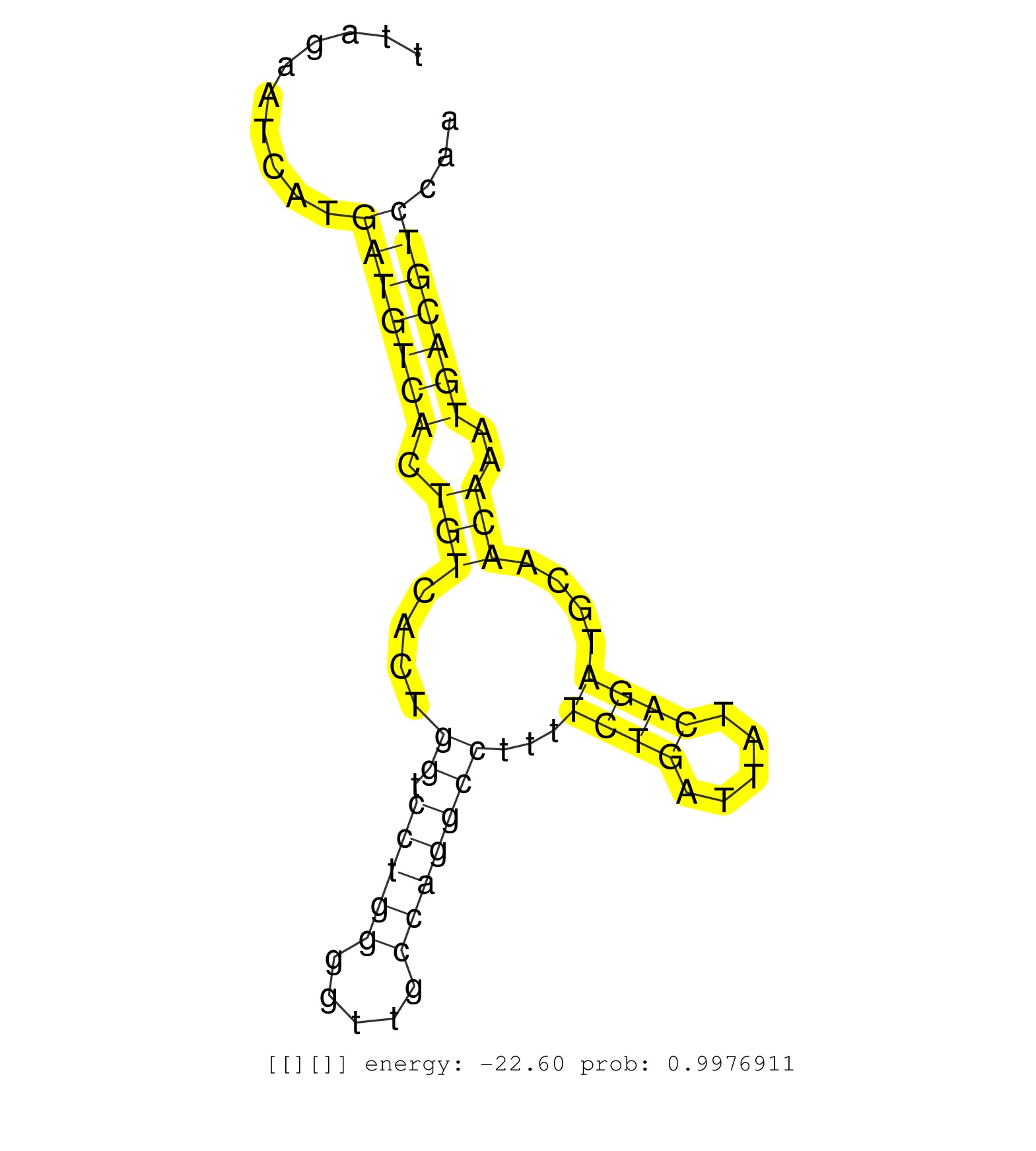
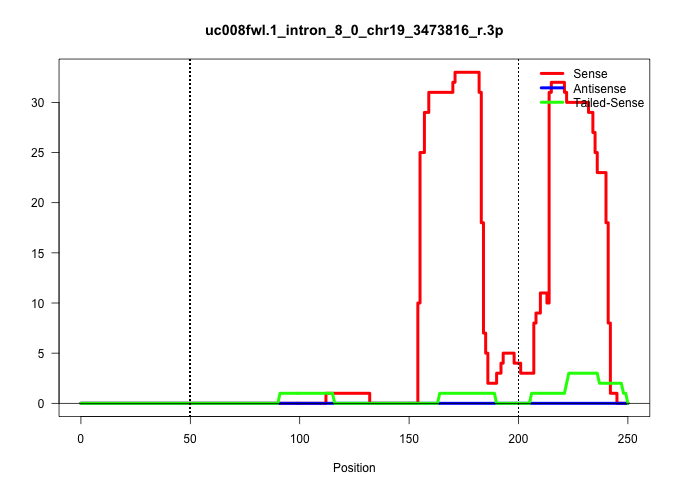
| Gene: Saps3 | ID: uc008fwl.1_intron_8_0_chr19_3473816_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(5) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(25) TESTES |
| ATTTTTCTTTTTCTAAAGAAAAACACACCAACCTTTAAAGTTCCATGGCTGTGTGAGTTAAAGTGGTTTTCTGGAACACAAATGGGCAAATAGTGTATAGAATGTCATTAGAATCATGATGTCACTGTCACTGGTCCTGGGGTTGCCAGGCCTTTTCTGATTATCAGATGCAACAAATGACGTCCAATTTTATTGACCAGTTTGGCTTCAACGATGAGAAGTTTGCAGACCAAGACGACATTGGCAAGTG .....................................................................................................................(((((((.(((....((.(((((.....)))))))...((((.....))))....)))..))))))).................................................................. ...........................................................................................................108............................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT1() Testes Data. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................TGAGAAGTTTGCAGACCAAGACGACAT......... | 27 | 1 | 10.00 | 10.00 | 2.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - |
| ......................................................................................................................................................................................................................TGAGAAGTTTGCAGACCAAGACGACA.......... | 26 | 1 | 8.00 | 8.00 | 3.00 | - | 1.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TCTGATTATCAGATGCAACAAATGACGT................................................................... | 28 | 1 | 8.00 | 8.00 | 3.00 | - | 1.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGAGAAGTTTGCAGACCAAGACGACATT........ | 28 | 1 | 6.00 | 6.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTCTGATTATCAGATGCAACAAATGACGTC.................................................................. | 30 | 1 | 5.00 | 5.00 | - | - | - | - | 2.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TCTGATTATCAGATGCAACAAATGACGTC.................................................................. | 29 | 1 | 5.00 | 5.00 | 2.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTCTGATTATCAGATGCAACAAATGACGT................................................................... | 29 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................AATCATGATGTCACTGTCACT...................................................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAACGATGAGAAGTTTGCAGACCAAG................ | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAACGATGAGAAGTTTGCAGACCAAGAC.............. | 29 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................ATTATCAGATGCAACAAATGACGTCCA................................................................ | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TATTGACCAGTTTGGCTTCAACG..................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGATTATCAGATGCAACAAATGACG.................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGATTATCAGATGCAACAAATGACGTCC................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGATTATCAGATGCAACAAATGACGTC.................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTGACCAGTTTGGCTTCAACGATGAGAAG............................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TCTGGAACACAAATGGGCAAA................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................................TGAGAAGTTTGCAGACCAAGACGACATTGGC..... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CAACGATGAGAAGTTTGCAGACCAAGA............... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TCTGATTATCAGATGCAACAAATGACGTCC................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TGACCAGTTTGGCTTCAACGATGAGAAGT............................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GAGAAGTTTGCAGACCAAGACGACATT........ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TCAGATGCAACAAATGACGTCCAATTTTATT........................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TTCAACGATGAGAAGTTTGCAGACCAAGttt............. | 31 | ttt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CAACAAATGACGTCCAATTTTATTGACC.................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGATTATCAGATGCAACAAATGACGTCCA................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TCTGATTATCAGATGCAACAAATGACG.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAACGATGAGAAGTTTGCAGACCAAGA............... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGCAGACCAAGACGACATTGGCAAtgt | 27 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................ACGATGAGAAGTTTGCAGACCAAGACGACA.......... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................................................................ACGATGAGAAGTTTGCAGACCA.................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................TGCAACAAATGACGTCCAATTTTATTGAC..................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................ATCATGATGTCACTGTCACT...................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................................................................AACAAATGACGTCCAATTTTATTGACCAGT................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CAGATGCAACAAATGACGTCCAATct............................................................ | 26 | ct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................AGTGTATAGAATGTCATTAGAATta...................................................................................................................................... | 25 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTGCAGACCAAGACGACATTGGCAAt.. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATTTTTCTTTTTCTAAAGAAAAACACACCAACCTTTAAAGTTCCATGGCTGTGTGAGTTAAAGTGGTTTTCTGGAACACAAATGGGCAAATAGTGTATAGAATGTCATTAGAATCATGATGTCACTGTCACTGGTCCTGGGGTTGCCAGGCCTTTTCTGATTATCAGATGCAACAAATGACGTCCAATTTTATTGACCAGTTTGGCTTCAACGATGAGAAGTTTGCAGACCAAGACGACATTGGCAAGTG .....................................................................................................................(((((((.(((....((.(((((.....)))))))...((((.....))))....)))..))))))).................................................................. ...........................................................................................................108............................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT1() Testes Data. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............AGAAAAACACACCAACCTTgaat....................................................................................................................................................................................................................... | 23 | gaat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |