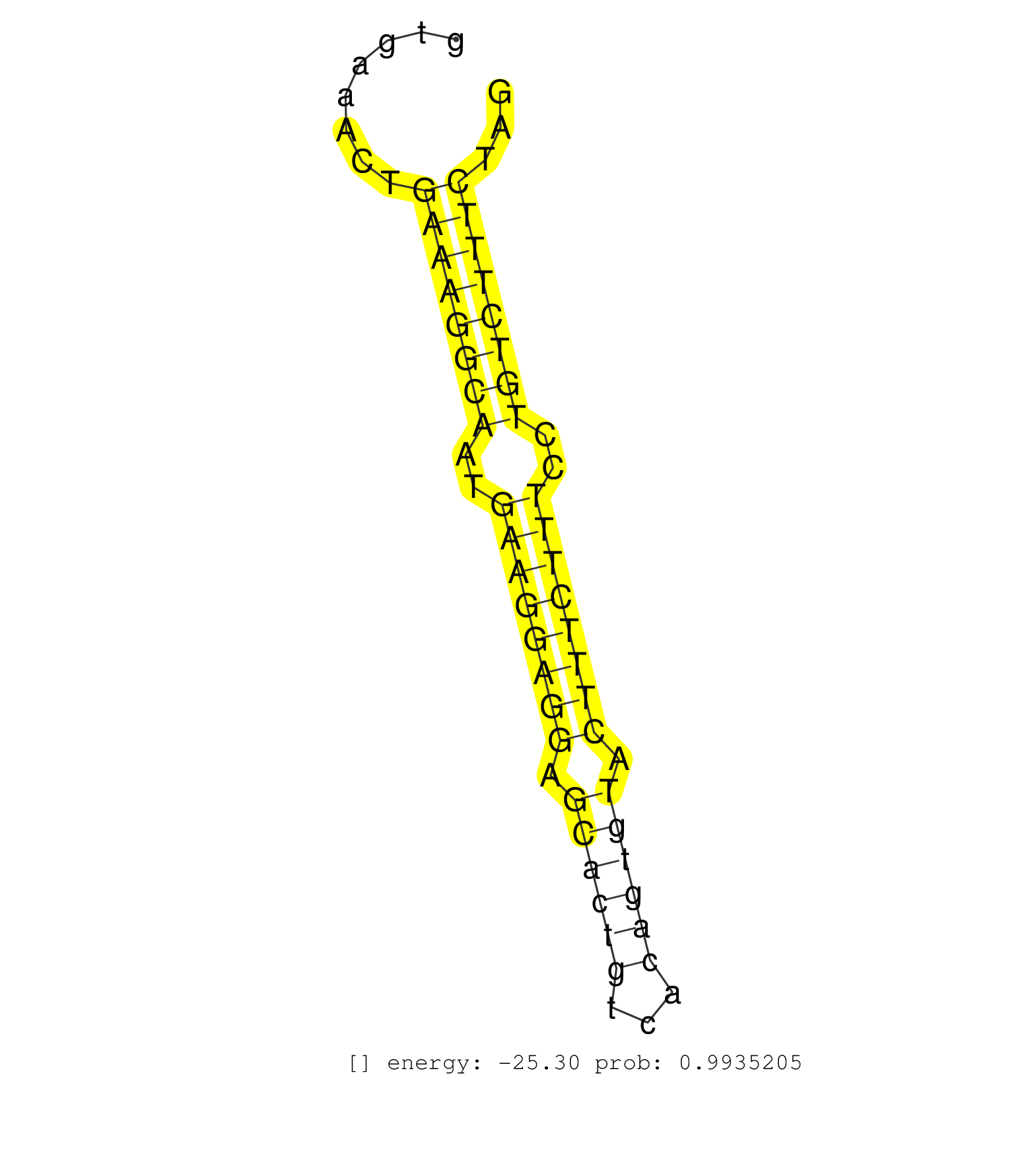

| Gene: Mrpl21 | ID: uc008fwd.1_intron_4_0_chr19_3287756_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| CTACACAGGAAGCAGAGGTAGGAGGGTCAGAGGTCAGTTTATTCCTTGGATGTAAAGAGAATTTAAGGTTGGTCTGGGCTGCATGAGACCCCACTGCAGAACCAGGAATTCTCTGGGGAGCTTCCGCAAGTTGTGCGTGAAACTGAAAGGCAATGAAGGAGGAGCACTGTCACAGTGTACTTTCTTTCCTGTCTTTCTAGAAAGGAGCTTGTACGAGTTGAAGCCACAGTCATTGAAAAGACAGAATCAT ................................................................................................................................................((((((((..((((((((.((((((...)))))).))))))))..))))))))..................................................... ........................................................................................................................................137............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................AAGGAGCTTGTACGAGTTGAAGCCACA...................... | 27 | 1 | 6.00 | 6.00 | - | - | 1.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................TGAAAGGCAATGAAGGAGGAG...................................................................................... | 21 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ACTGAAAGGCAATGAAGGAGGAGa..................................................................................... | 24 | a | 3.00 | 1.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ACTGAAAGGCAATGAAGGAGGAGC..................................................................................... | 24 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ACTGAAAGGCAATGAAGGAGGA....................................................................................... | 22 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGAAAGGCAATGAAGGAGGAGCAa................................................................................... | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TTGTACGAGTTGAAGCCACAGTCATTG............... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............AGAGGTAGGAGGGTtcat........................................................................................................................................................................................................................... | 18 | tcat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAACTGAAAGGCAATGAAGGAGGAGC..................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................GAAAGGCAATGAAGGAGGAGC..................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................ACTGAAAGGCAATGAAGGAGGAGCA.................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....CAGGAAGCAGAGGTAGatgt................................................................................................................................................................................................................................. | 20 | atgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................................................................AAGGAGCTTGTACGAGTTGAAGCCAC....................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ACTGAAAGGCAATGAAGGAGGAG...................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ACTGAAAGGCAATGAAGGAGG........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGAAAGGCAATGAAGGAGGAGC..................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................CAAGTTGTGCGTGAcgc........................................................................................................... | 17 | cgc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................GCAATGAAGGAGGAGCggg.................................................................................. | 19 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................AATTTAAGGTTGGTaacc............................................................................................................................................................................. | 18 | aacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................TACTTTCTTTCCTGTCTTTCTAGA................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AGGAGCTTGTACGAGTTGAAGCCACAGTC................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTACACAGGAAGCAGAGGTAGGAGGGTCAGAGGTCAGTTTATTCCTTGGATGTAAAGAGAATTTAAGGTTGGTCTGGGCTGCATGAGACCCCACTGCAGAACCAGGAATTCTCTGGGGAGCTTCCGCAAGTTGTGCGTGAAACTGAAAGGCAATGAAGGAGGAGCACTGTCACAGTGTACTTTCTTTCCTGTCTTTCTAGAAAGGAGCTTGTACGAGTTGAAGCCACAGTCATTGAAAAGACAGAATCAT ................................................................................................................................................((((((((..((((((((.((((((...)))))).))))))))..))))))))..................................................... ........................................................................................................................................137............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................TGTACTTTCTTTCCTGTCTTTCTAGAAA............................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................GTACTTTCTTTCCTGTCTTTCTAGAAA............................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |