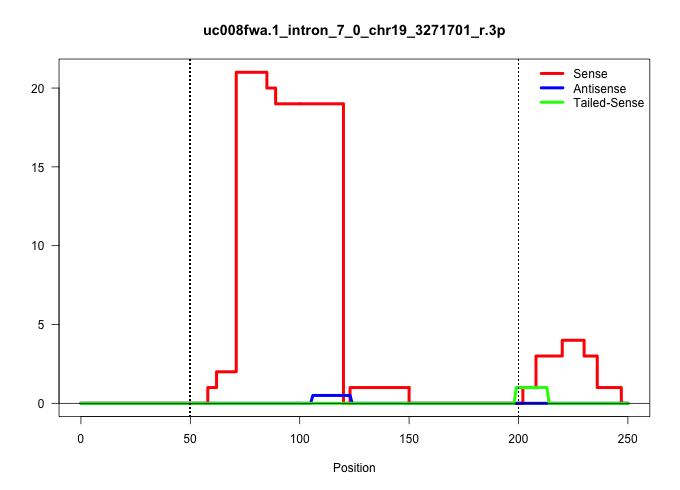
| Gene: Ighmbp2 | ID: uc008fwa.1_intron_7_0_chr19_3271701_r.3p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(8) TESTES |
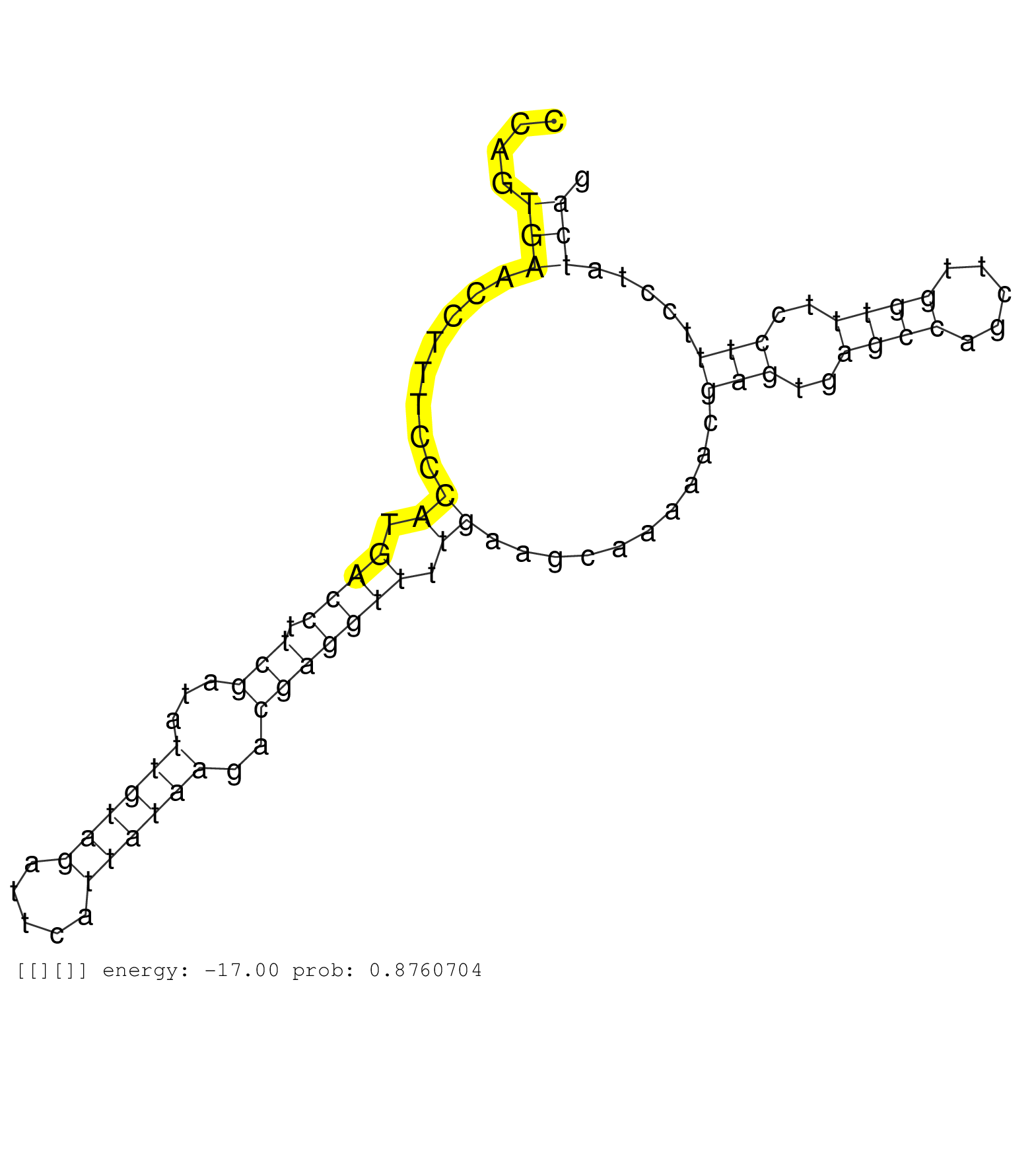
| GAGGTCAGAGGACAGCTTGGAGAAGGCAGTTCTGTTCTTCCAACATGTGGGCCTTAGGACTTTAACTCAGGTCATTAGGTGTGGTGGCAAGCTCCTTTATCCAGTGAACCTTTCCCATGACCTTCGATATTGTAGATTCATTATAAGACGAGGTTTTGAAGCAAAAACGAGTGAGCCAGCTTGGTTTCCTTTCCTATCAGGTGCATCGTCTGATGGCCCTCTGAAGCTGCTGCCTGAGGACTACTTTGAT ........................................................................................................(((........((.((((.(((...((((((.....))))))..))))))).))..........(((..((((.....))))..))).....)))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................TCATTAGGTGTGGTGGCAAGCTCCTTTATCCAGTGAACCTTTCCCATGACCT............................................................................................................................... | 52 | 1 | 18.00 | 18.00 | 18.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TCTGATGGCCCTCTGAAGCTGCTGCCTG.............. | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - |
| .......................................................................................................................................................................................................GGTGCATCGTCgacc.................................... | 15 | gacc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - |
| ..............................................................TAACTCAGGTCATTAGGTGTGGTGGCA................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................TCGATATTGTAGATTCATTATAAGACG.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CTGAAGCTGCTGCCTGAGGACTACTTT... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................GCATCGTCTGATGGCCCTCTGAAGCTGC.................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ..........................................................ACTTTAACTCAGGTCATTAGGTGTGGT..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| GAGGTCAGAGGACAGCTTGGAGAAGGCAGTTCTGTTCTTCCAACATGTGGGCCTTAGGACTTTAACTCAGGTCATTAGGTGTGGTGGCAAGCTCCTTTATCCAGTGAACCTTTCCCATGACCTTCGATATTGTAGATTCATTATAAGACGAGGTTTTGAAGCAAAAACGAGTGAGCCAGCTTGGTTTCCTTTCCTATCAGGTGCATCGTCTGATGGCCCTCTGAAGCTGCTGCCTGAGGACTACTTTGAT ........................................................................................................(((........((.((((.(((...((((((.....))))))..))))))).))..........(((..((((.....))))..))).....)))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................AACCTTTCCCATGACCTT.............................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 |