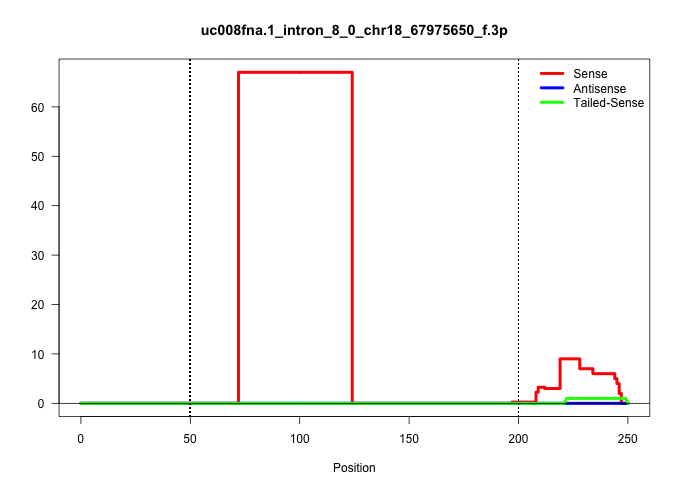
| Gene: AK158414 | ID: uc008fna.1_intron_8_0_chr18_67975650_f.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(10) TESTES |
| TTCTCTGTATGCTCCCGGTCTTCCTATGTCCATCCCATGTCCATCCCATGTGTCCATCCCATGTCCATCCCGTATGATGCTTGTTTTCCCTTGCTTCCAACCTGCAGAATACGTCAGATGGATACAGCTAAGAAGACAGCTACTGCATTAGACTACCTAGACTTTGACATTGCTCCATTAACCACTTCTCCTTATTTTAGGTGGCTGTGTAGGTACTGTCGATAATAAAATGCAGGTAGATAATGTGATG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................TATGATGCTTGTTTTCCCTTGCTTCCAACCTGCAGAATACGTCAGATGGATA.............................................................................................................................. | 52 | 1 | 67.00 | 67.00 | 67.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GTAGGTACTGTCGATAATAA...................... | 20 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CGATAATAAAATGCAGGTAGATAATGTG... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................................................................................CGATAATAAAATGCAGGTAGATAATGT.... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................................CGATAATAAAATGCAGGTAGATAATG..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CGATAATAAAATGCAGGTAGATAAT...... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TAGGTACTGTCGATAATAAAATGCA................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................................TAATAAAATGCAGGTAGATAATGTGAaa | 28 | aa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................TAGGTGGCTGTGTAG...................................... | 15 | 8 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | 0.25 |
| TTCTCTGTATGCTCCCGGTCTTCCTATGTCCATCCCATGTCCATCCCATGTGTCCATCCCATGTCCATCCCGTATGATGCTTGTTTTCCCTTGCTTCCAACCTGCAGAATACGTCAGATGGATACAGCTAAGAAGACAGCTACTGCATTAGACTACCTAGACTTTGACATTGCTCCATTAACCACTTCTCCTTATTTTAGGTGGCTGTGTAGGTACTGTCGATAATAAAATGCAGGTAGATAATGTGATG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|