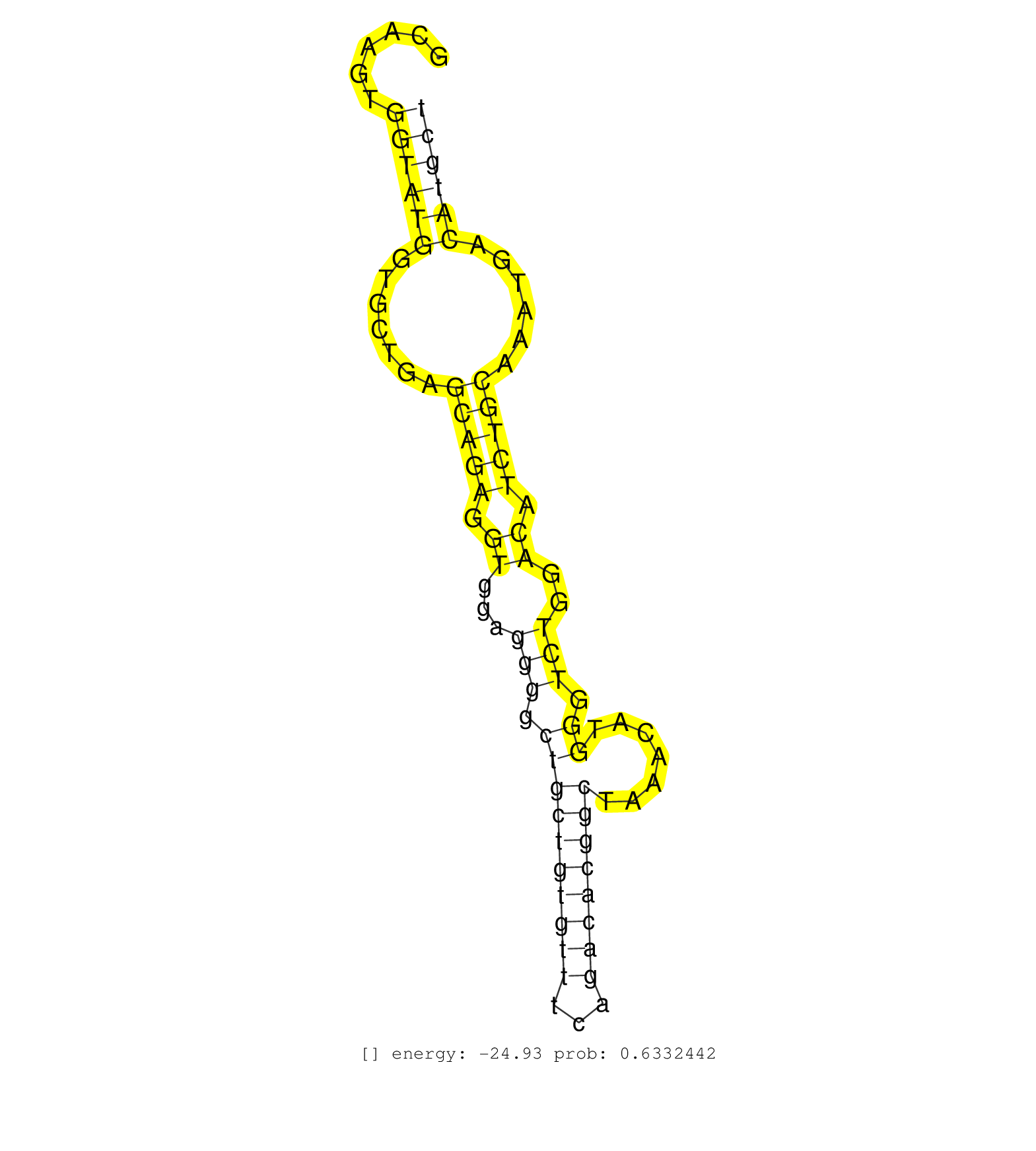
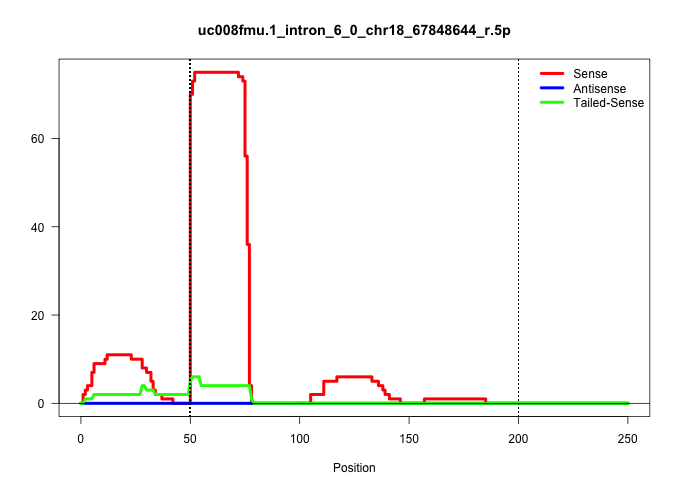
| Gene: Ptpn2 | ID: uc008fmu.1_intron_6_0_chr18_67848644_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
| CCAGCTTAGTTGACATAGAAGAGGCACAAAGAAGTTACATCTTAACACAGGCAAGTGGTATGGTGCTGAGCAGAGGTGGAGGGGCTGCTGTGTTTCAGACACGGCTAAACATGGGTCTGGACATCTGCAAATGACATGCTATCTTTTAGTTTTCTGATTCTGAAATCAGTTTTGCAAATGCCATAGTTTTCAGATTTCGAATATATTGATAAAAAGTAAAATTGTTCCTAATGTTACTATTTTCAATGGA ........................................................((((((.......(((((.((...(((.((((((((((...)))))))).......)).)))..)).)))))......)))))).............................................................................................................. ..................................................51.......................................................................................140............................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GCAAGTGGTATGGTGCTGAGCAGAGGT............................................................................................................................................................................. | 27 | 1 | 30.00 | 30.00 | 10.00 | 5.00 | 6.00 | - | 5.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTATGGTGCTGAGCAGAGG.............................................................................................................................................................................. | 26 | 1 | 19.00 | 19.00 | 6.00 | 5.00 | 1.00 | 1.00 | 1.00 | 2.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTATGGTGCTGAGCAGAG............................................................................................................................................................................... | 25 | 1 | 17.00 | 17.00 | 6.00 | 8.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ....................................................AAGTGGTATGGTGCTGAGCAGAGGT............................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................AAGAAGTTACATCTTAACACAGGgccc................................................................................................................................................................................................... | 27 | gccc | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ..................................................GCAAGTGGTATGGTGCTGAGCAGAGGTG............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....TTAGTTGACATAGAAGAGGCACAAAGAA......................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CAGCTTAGTTGACATAGAAGAGGCACA.............................................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GCAAGTGGTATGGTGCTGAGCAGAGGTt............................................................................................................................................................................ | 28 | t | 2.00 | 30.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TCTGAAATCAGTTTTGCAAATG...................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ...................................................CAAGTGGTATGGTGCTGAGCAGAGGTG............................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAAACATGGGTCTGGACATCTGCAAATGACA.................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TTCTGAAATCAGTTTTGCAAATGCCATA................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................CAAGTGGTATGGTGCTGAGCAGAGG.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTATGGTGCTGAGCAGA................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...GCTTAGTTGACATAGAAGAGGCACAAA............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TAGTTGACATAGAAGAGGCACAAAGAAt........................................................................................................................................................................................................................ | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAAACATGGGTCTGGACATCTGCAAATG..................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGTCTGGACATCTGCAAATGACATG................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................CAAGTGGTATGGTGCTGAGCAGAGGTGt........................................................................................................................................................................... | 28 | t | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTATGGTGCTGAGCA.................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....TTAGTTGACATAGAAGAGGCACAAAGA.......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGTCTGGACATCTGCAAATGACATGC............................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..AGCTTAGTTGACATAGAAGAGGCACcaa............................................................................................................................................................................................................................ | 28 | caa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTATGGTGCTGAGCAGAGGgg............................................................................................................................................................................ | 28 | gg | 1.00 | 19.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..AGCTTAGTTGACATAGAAGAG................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGTCTGGACATCTGCAAATGACATGCTA............................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGGACATCTGCAAATGACATGCTATCTTT........................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......TAGTTGACATAGAAGAGGCACAAAGA.......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TAGTTGACATAGAAGAGGCACAAAGAAG........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............ACATAGAAGAGGCACAAAGAAGTTACATCT................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........GACATAGAAGAGGCACAAAGAAGTTA..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GCTGAGCAGAGGTGGAG......................................................................................................................................................................... | 17 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 |
| CCAGCTTAGTTGACATAGAAGAGGCACAAAGAAGTTACATCTTAACACAGGCAAGTGGTATGGTGCTGAGCAGAGGTGGAGGGGCTGCTGTGTTTCAGACACGGCTAAACATGGGTCTGGACATCTGCAAATGACATGCTATCTTTTAGTTTTCTGATTCTGAAATCAGTTTTGCAAATGCCATAGTTTTCAGATTTCGAATATATTGATAAAAAGTAAAATTGTTCCTAATGTTACTATTTTCAATGGA ........................................................((((((.......(((((.((...(((.((((((((((...)))))))).......)).)))..)).)))))......)))))).............................................................................................................. ..................................................51.......................................................................................140............................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|