
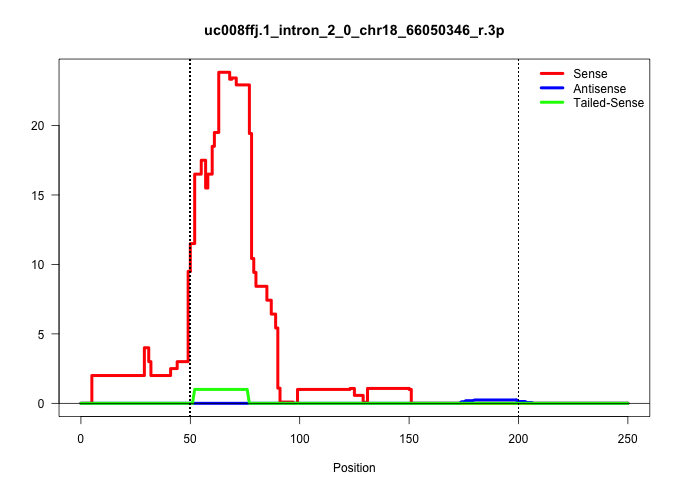
| Gene: BC053977 | ID: uc008ffj.1_intron_2_0_chr18_66050346_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(4) PIWI.mut |
(23) TESTES |
| ACTGCTGCTACTTGCCGCCTAGTTGTTCCTGAGCTGACACGTGGTCACCTGAAACTGGACTCCTAGGATGATTTGGCAGGAATGGGCCCCTCCCCCTTCTTCATAACCCGGTGTCTCTGAAGAGTAAAATTGAACCTTGATCAGACTGACGCCTTGGTTCCATTCTTTCTCGCGCCGCCTAGTTCCCTCTTCTCTTCCAGATTCCAAGATGGCTCCCAGGCTAGAACCCAGACATGTGGGCCGCTGGCCA ......................................................((((....))))...........(((((.(((......))).)))))........(((((((((((.(((...........)))..)))))..))))))................................................................................................. ..................................................51....................................................................................................153............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................TGGACTCCTAGGATGATTTGGCAGGA......................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | - | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AACTGGACTCCTAGGATGATTTGGCA............................................................................................................................................................................ | 26 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................TGAAACTGGACTCCTAGGATGATTTGGC............................................................................................................................................................................. | 28 | 2 | 3.50 | 3.50 | 1.00 | - | - | 1.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ...............................................................TAGGATGATTTGGCAGGAATGGGCCCC................................................................................................................................................................ | 27 | 3 | 3.33 | 3.33 | 1.00 | - | - | - | - | 0.67 | - | - | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | 0.33 | 0.67 | - | - | - |
| .................................................TGAAACTGGACTCCTAGGATGATTTGGCA............................................................................................................................................................................ | 29 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................AAATTGAACCTTGATCAGAaaaa..................................................................................................... | 23 | aaaa | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGGACTCCTAGGATGATTTGGCAGG.......................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TGAGCTGACACGTGGTCACCTGAAACTG................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GAAACTGGACTCCTAGGATGATTTGGCA............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAAAATTGAACCTTGATCAGACTGACGC.................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................TGATTTGGCAGGAATGGGCCgctc.............................................................................................................................................................. | 24 | gctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGCTACTTGCCGCCTAGTTGTTCCTG........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CCTAGGATGATTTGGCAGGAATGGGCCCC................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TAGGATGATTTGGCAGGAATGGGCCCCT............................................................................................................................................................... | 28 | 3 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TCCTAGGATGATTTGGCAGGAATGGGCCC................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AACTGGACTCCTAGGATGATTTGtc............................................................................................................................................................................. | 25 | tc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TCCTAGGATGATTTGGCAGGAATGGGC................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................ACTCCTAGGATGATTTGGCAGGAATGG..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GAACCTTGATCAGACTGACG................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................AACTGGACTCCTAGGATGATTTGGCAG........................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TCTTCTCTTCCAGATTCCAAGATccct.................................... | 27 | ccct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TCACCTGAAACTGGACTCCTAGGgtg.................................................................................................................................................................................... | 26 | gtg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....TGCTACTTGCCGCCTAGTTGTTCCTGA.......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GAAACTGGACTCCTAGGATGATTTGGC............................................................................................................................................................................. | 27 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TCACCTGAAACTGGACTCCTAGGATGA................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .........................................TGGTCACCTGAAACTGGACTCCTAGGA...................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTCATAACCCGGTGTCTCTGAAGAGTA............................................................................................................................ | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTCATAACCCGGTGTCTCTGAAGAGTAAAA......................................................................................................................... | 30 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTCATAACCCGGTGTCTCTGAAGAGT............................................................................................................................. | 26 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GATTTGGCAGGAATGGGCCCCTCCCCCT......................................................................................................................................................... | 28 | 11 | 0.09 | 0.09 | - | - | - | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................GTAAAATTGAACCTTGATCAGACTGAC.................................................................................................... | 27 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - | - |
| ACTGCTGCTACTTGCCGCCTAGTTGTTCCTGAGCTGACACGTGGTCACCTGAAACTGGACTCCTAGGATGATTTGGCAGGAATGGGCCCCTCCCCCTTCTTCATAACCCGGTGTCTCTGAAGAGTAAAATTGAACCTTGATCAGACTGACGCCTTGGTTCCATTCTTTCTCGCGCCGCCTAGTTCCCTCTTCTCTTCCAGATTCCAAGATGGCTCCCAGGCTAGAACCCAGACATGTGGGCCGCTGGCCA ......................................................((((....))))...........(((((.(((......))).)))))........(((((((((((.(((...........)))..)))))..))))))................................................................................................. ..................................................51....................................................................................................153............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....CTGCTACTTGCCGCCTAGTTGTTCCTG........................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CTGCTGCTACTTGCCGCCTA..................................................................................................................................................................................................................................... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TTCCCTCTTCTCTTCCAGATTCCAAGAt......................................... | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTTCTCGCGCCGagaa......................................................................... | 16 | agaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....TGCTACTTGCCGCCTAGTTGTTCCTGa........................................................................................................................................................................................................................... | 27 | a | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CCCTCTTCTCTTCCAGctgg.................................................. | 20 | ctgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGATCAGACTGACGCggca.................................................................................................. | 19 | ggca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................CCGCCTAGTTCCCTCTTCTCTTCCAGA................................................. | 27 | 7 | 1.00 | 1.00 | - | 0.71 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.29 | - | - |
| .............CGCCTAGTTGTTCCTGAGCTGACAtc................................................................................................................................................................................................................... | 26 | tc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CCTCTTCTCTTCCAGATTCCAAGAt......................................... | 25 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTCCCTCTTCTCTTCCAaaaa................................................... | 21 | aaaa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CCAGATTCCAAGATGGCTCCCAGGCTA........................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CTTTCTCGCGCCGCCTAGTTCCCTC............................................................. | 25 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ..............................................................................................................................................................................CCGCCTAGTTCCCTCTTCTCTTCCAG.................................................. | 26 | 9 | 0.22 | 0.22 | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| ............................................................................................................................................................................CGCCGCCTAGTTCCCTCTTCTCTTCCAG.................................................. | 28 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................CCGCCTAGTTCCCTCTTCTCTTCCAGATTC.............................................. | 30 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................GCCTAGTTCCCTCTTCTCTTCCAGATTC.............................................. | 28 | 13 | 0.08 | 0.08 | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AGTTCCCTCTTCTCTTCCAGATTCCAAga........................................... | 29 | ga | 0.05 | 0.00 | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................AGTTCCCTCTTCTCTTCCAGATTCCAA........................................... | 27 | 19 | 0.05 | 0.05 | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |