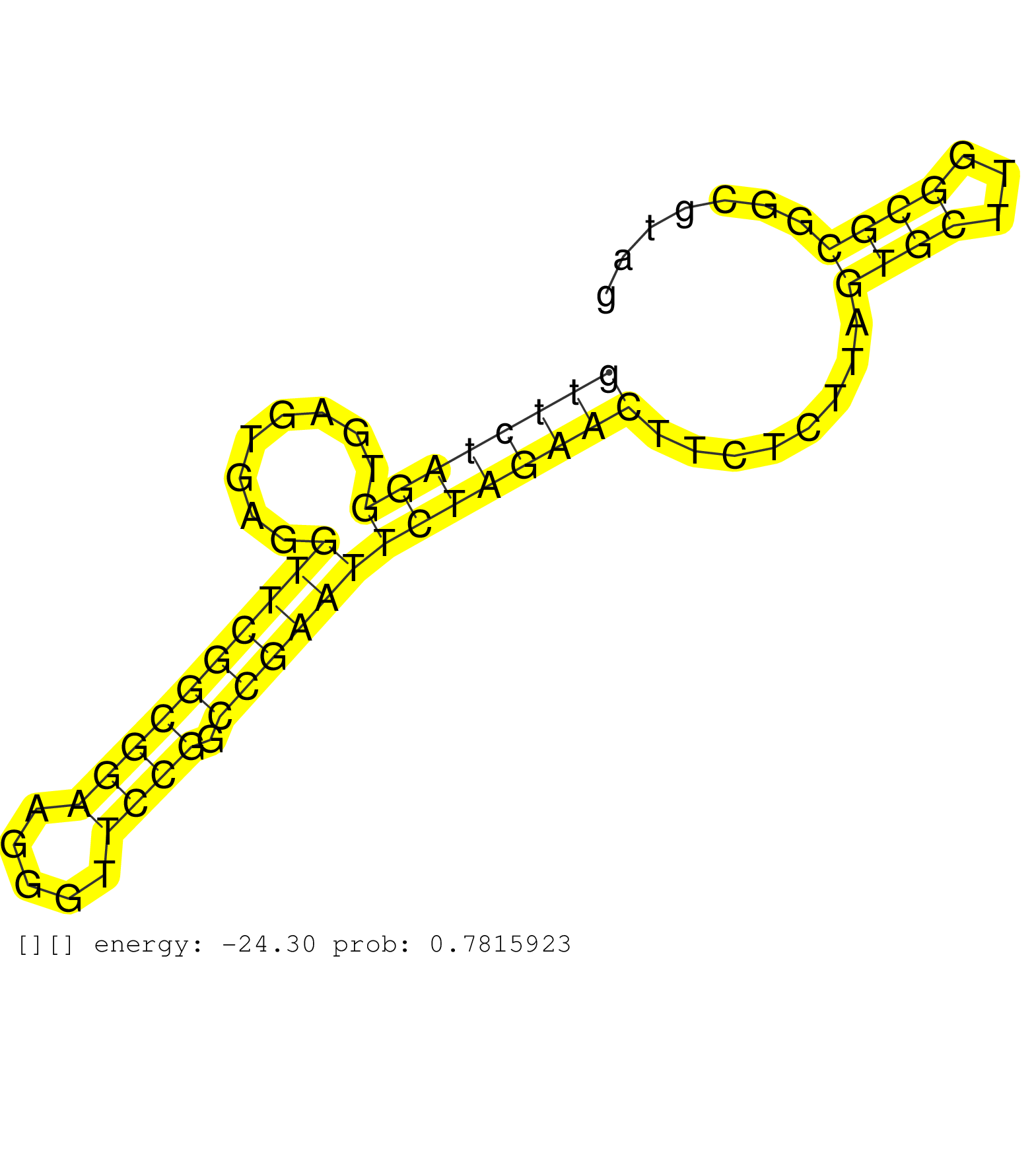
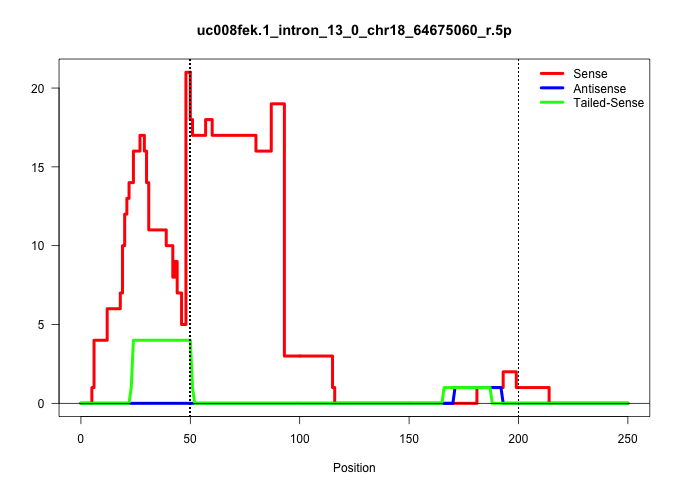
| Gene: Nars | ID: uc008fek.1_intron_13_0_chr18_64675060_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| AGTGTCTATGTCCTCGGAGGTGATTAGGGGGACTGCAGAGATGGTTCTAGGTGAGTGAGGTTCGGCGGAAGGGTTCCGGCCGAATTCTAGAACTTCTCTTAGTGCTTGGCGCGGCGTAGGCGCACTCCCAAATATTCGTGGGGCCCTGGTCGCTCGCCCCAGACCCTCAGACCTGGGCCACCTGGACACCGACTCGGTGGTCCGGGTCTGGTTTTGCTCCAGCCCTGTGCGTCCCTCGGCCACCGCCTCC ...........................................((((((((........((((((((((.....)))).))))))))))))))........((((...)))).......................................................................................................................................... ...........................................44.........................................................................119................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................AGGTGAGTGAGGTTCGGCGGAAGGGTTCCGGCCGAATTCTAGAACTTCTCTT...................................................................................................................................................... | 52 | 1 | 16.00 | 16.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TAGAACTTCTCTTAGTGCTTGGCGCGGC....................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......TATGTCCTCGGAGGTGATTAGGGGG........................................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - |
| ........................TAGGGGGACTGCAGAGATGGTTCTAGa....................................................................................................................................................................................................... | 27 | a | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TAGGGGGACTGCAGAGATGGTTCTAG........................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GTTCTAGGTGAGTGAGG.............................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CTCGGAGGTGATTAGGG............................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TCGGTGGTCCGGGTCTGGTTT.................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GGTGATTAGGGGGACTGCAGAGATGG.............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................GGGGACTGCAGAGATGGTTCTAGG....................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TAGGGGGACTGCAGAGATGGTTCTAGc....................................................................................................................................................................................................... | 27 | c | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................TAGAACTTCTCTTAGTGCTTGGCGCGGCG...................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................ATTAGGGGGACTGCAGAGATGGTTCTAG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................ATTAGGGGGACTGCAGA................................................................................................................................................................................................................... | 17 | 2 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CGGAGGTGATTAGGGGGACTGCAG.................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................AGGGGGACTGCAGAGATGGT............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GTGATTAGGGGGACTGCAGAGAT................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CTATGTCCTCGGAGGTGATTAGGGG............................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................TAGGGGGACTGCAGAGATGGTTCTA......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................GTGATTAGGGGGACTGCAGA................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TTAGGGGGACTGCAGAGATGGTTCTAGca...................................................................................................................................................................................................... | 29 | ca | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .GTGTCTATGTCCTCGGAGG...................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TCAGACCTGGGCCACCTGGAaa.............................................................. | 22 | aa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGGTTCGGCGGAAGGGTTCCGGC.......................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGATTAGGGGGACTGCAGAGAT................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................GTGATTAGGGGGACTGCAGAGATGGTT............................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GATTAGGGGGACTGCAGAGATGG.............................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGATTAGGGGGACTGCAGAGATGGTT............................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CTGGACACCGACTCGGTG................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CTCGGAGGTGATTAGGGG............................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGTGTCTATGTCCTCGGAGGTGATTAGGGGGACTGCAGAGATGGTTCTAGGTGAGTGAGGTTCGGCGGAAGGGTTCCGGCCGAATTCTAGAACTTCTCTTAGTGCTTGGCGCGGCGTAGGCGCACTCCCAAATATTCGTGGGGCCCTGGTCGCTCGCCCCAGACCCTCAGACCTGGGCCACCTGGACACCGACTCGGTGGTCCGGGTCTGGTTTTGCTCCAGCCCTGTGCGTCCCTCGGCCACCGCCTCC ...........................................((((((((........((((((((((.....)))).))))))))))))))........((((...)))).......................................................................................................................................... ...........................................44.........................................................................119................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................CCTGGGCCACCTGGACACCGAC......................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCCAGCCCTGTGCGTagg.................. | 18 | agg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................................................TTTTGCTCCAGCCCTaa........................ | 17 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |