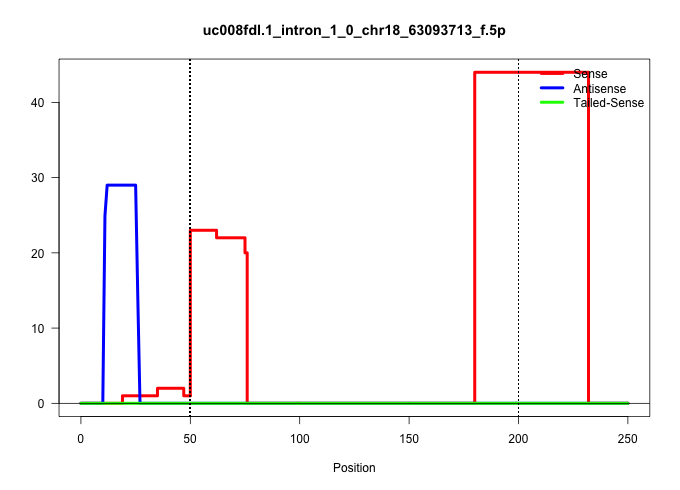
| Gene: Apcdd1 | ID: uc008fdl.1_intron_1_0_chr18_63093713_f.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(10) TESTES |
| ATCACAGTGCAGATGCCCCCGACCATCGAGGGCCACTGGGTGTCCACAGGGTAAGAGCTGCATTTGAGACGGGAAGTGCAGAGAGAGCAACCTGGGAAGCAGGGCCTGAGGAAGACCCTCTGATGGGTTTAGATGCTAGCTTTGCACAATCAACAGCAAGAGAGAGAACCAGGCATCTCTTGGAAATTATGGTTGGTTATGCAAGCAGAACTAAGAGCTGCAGAAATCTTACTCCAAAGCAGCAGCGGTA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................TGGAAATTATGGTTGGTTATGCAAGCAGAACTAAGAGCTGCAGAAATCTTAC.................. | 52 | 1 | 44.00 | 44.00 | 44.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAGCTGCATTTGAGACGGGAAG.............................................................................................................................................................................. | 26 | 1 | 20.00 | 20.00 | - | - | 15.00 | - | 5.00 | - | - | - | - | - |
| ..................................................GTAAGAGCTGCATTTGAGACGGGAA............................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ...................CGACCATCGAGGGCCACTGGGTGTCCAC........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................CTGGGTGTCCACAGGGTAAGAGCTGCA............................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ATCACAGTGCAGATGCCCCCGACCATCGAGGGCCACTGGGTGTCCACAGGGTAAGAGCTGCATTTGAGACGGGAAGTGCAGAGAGAGCAACCTGGGAAGCAGGGCCTGAGGAAGACCCTCTGATGGGTTTAGATGCTAGCTTTGCACAATCAACAGCAAGAGAGAGAACCAGGCATCTCTTGGAAATTATGGTTGGTTATGCAAGCAGAACTAAGAGCTGCAGAAATCTTACTCCAAAGCAGCAGCGGTA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........GATGCCCCCGACCAT................................................................................................................................................................................................................................ | 15 | 1 | 15.00 | 15.00 | - | 15.00 | - | - | - | - | - | - | - | - |
| ...........GATGCCCCCGACCATC............................................................................................................................................................................................................................... | 16 | 1 | 10.00 | 10.00 | - | 10.00 | - | - | - | - | - | - | - | - |
| ........GATGCCCCCGACagt................................................................................................................................................................................................................................... | 15 | agt | 6.00 | 0.00 | - | - | - | 6.00 | - | - | - | - | - | - |
| ............ATGCCCCCGACCATC............................................................................................................................................................................................................................... | 15 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - |
| ..........GATGCCCCCGACCATa................................................................................................................................................................................................................................ | 16 | a | 4.00 | 0.00 | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - |
| .......GATGCCCCCGACagta................................................................................................................................................................................................................................... | 16 | agta | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - |
| .......GATGCCCCCGACCagta.................................................................................................................................................................................................................................. | 17 | agta | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ........GATGCCCCCGACCATCagt............................................................................................................................................................................................................................... | 19 | agt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - |