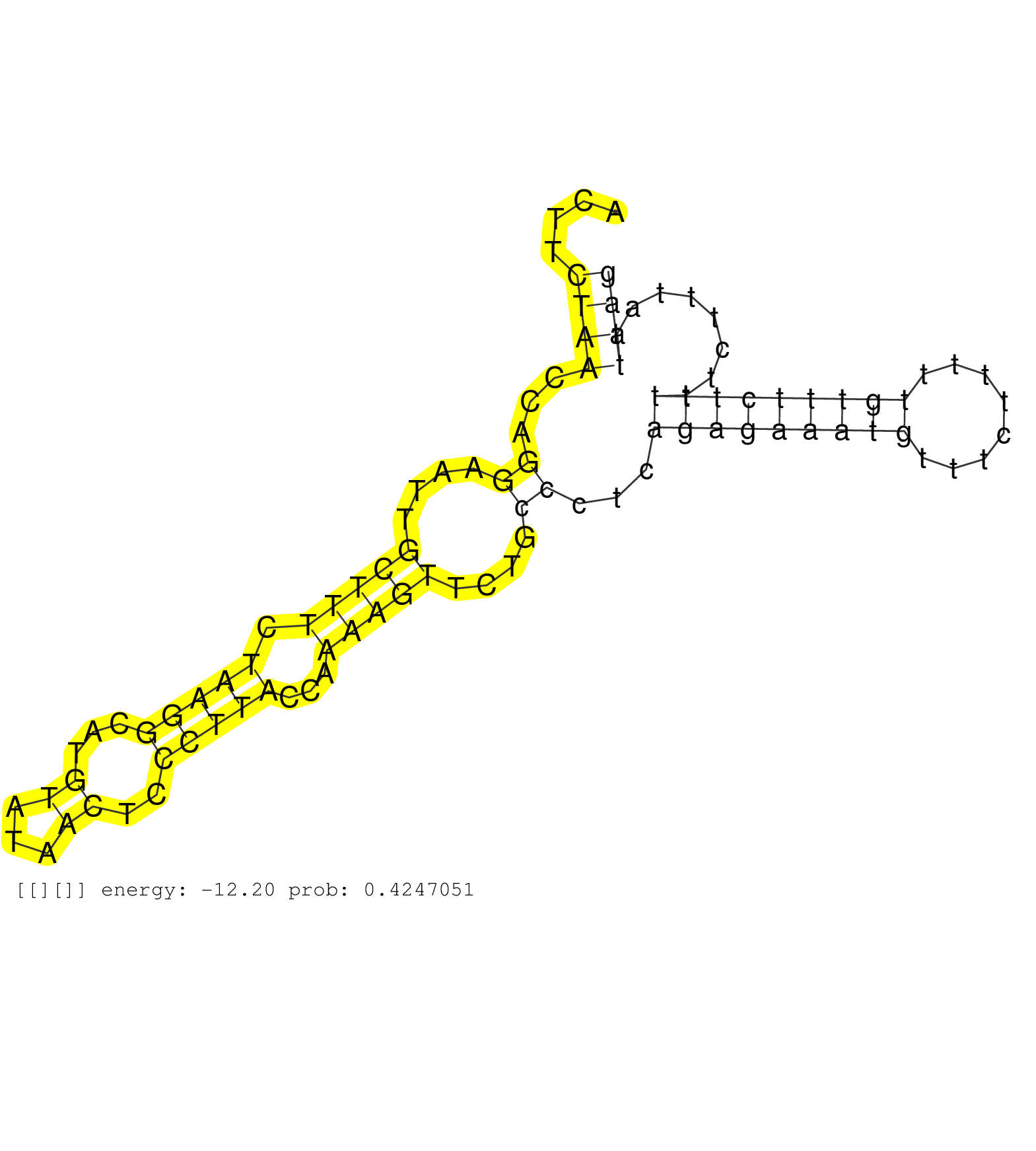

| Gene: Fbxo38 | ID: uc008fde.1_intron_7_0_chr18_62686925_r.3p | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(2) PIWI.ip |
(20) TESTES |
| AGATGCTCCAAGGATGTTTCCAGAATGAGTATCTGTTTTGTGGAGTAACCGCTAGGAATAATTCACCTTGAGGTAATTTGATGGTAGCCGCAAAGCATTTACTTCTAACCAGGAATTGCTTTCTAAGGCATGTATAACTCCCTTACCAAAAGTTCTGCCCTCAGAGAAATGTTTCTTTTTGTTTCTTTTTCTTTAATTAGGTGGTTTTAGGAACTTGCACACTATTGTCTTAGGAGCTTGCAAGAATGCC ........................................................................................................((((...((....(((((.(((((...((...))..)))))...)))))....))...(((((((((........)))))))))........)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................ACCTTGAGGTAATTTGATGGTAGCCGCAAAGCATTTACTTCTAACCAGGAAT...................................................................................................................................... | 52 | 1 | 74.00 | 74.00 | 74.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TAGGAACTTGCACACTATTGTCTTAGGAGC............. | 30 | 1 | 15.00 | 15.00 | - | 7.00 | - | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................................TAGGAACTTGCACACTATTGTCTTA.................. | 25 | 1 | 5.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................TAGGAACTTGCACACTATTGTCTTAGGAG.............. | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................TAGGAACTTGCACACTATTGTCTTAGG................ | 27 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TAGGAACTTGCACAgatc......................... | 18 | gatc | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AACTTGCACACTATTGTCTTAGt................ | 23 | t | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TTGCACACTATTGTCTTAGGAGCTTGC......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................ACCGCTAGGAATAATggaa........................................................................................................................................................................................ | 19 | ggaa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TATTGTCTTAGGAGCTTGC......... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGGTTTTAGGAACTTGCACACcat......................... | 24 | cat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGCACACTATTGTCTTAGGAGCTTGC......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TAGGAACTTGCACACTATTGTCTTAGtagc............. | 30 | tagc | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................AATTAGGTGGTTTTAGGAACTT.................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................................CTATTGTCTTAGGAGCTTGCAAGAATG.. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................................................................GGTGGTTTTAGGAACTgagg............................... | 20 | gagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TAGGAACTTGCACACTATTGTCTTAG................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................AGTATCTGTTTTGTGGgttt........................................................................................................................................................................................................... | 20 | gttt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TAGGAACTTGCACACTATTGTCTTAGGA............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TTAGGAGCTTGCAAG...... | 15 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGATGCTCCAAGGATGTTTCCAGAATGAGTATCTGTTTTGTGGAGTAACCGCTAGGAATAATTCACCTTGAGGTAATTTGATGGTAGCCGCAAAGCATTTACTTCTAACCAGGAATTGCTTTCTAAGGCATGTATAACTCCCTTACCAAAAGTTCTGCCCTCAGAGAAATGTTTCTTTTTGTTTCTTTTTCTTTAATTAGGTGGTTTTAGGAACTTGCACACTATTGTCTTAGGAGCTTGCAAGAATGCC ........................................................................................................((((...((....(((((.(((((...((...))..)))))...)))))....))...(((((((((........)))))))))........)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....TCCAAGGATGTTTCCAat.................................................................................................................................................................................................................................... | 18 | at | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |