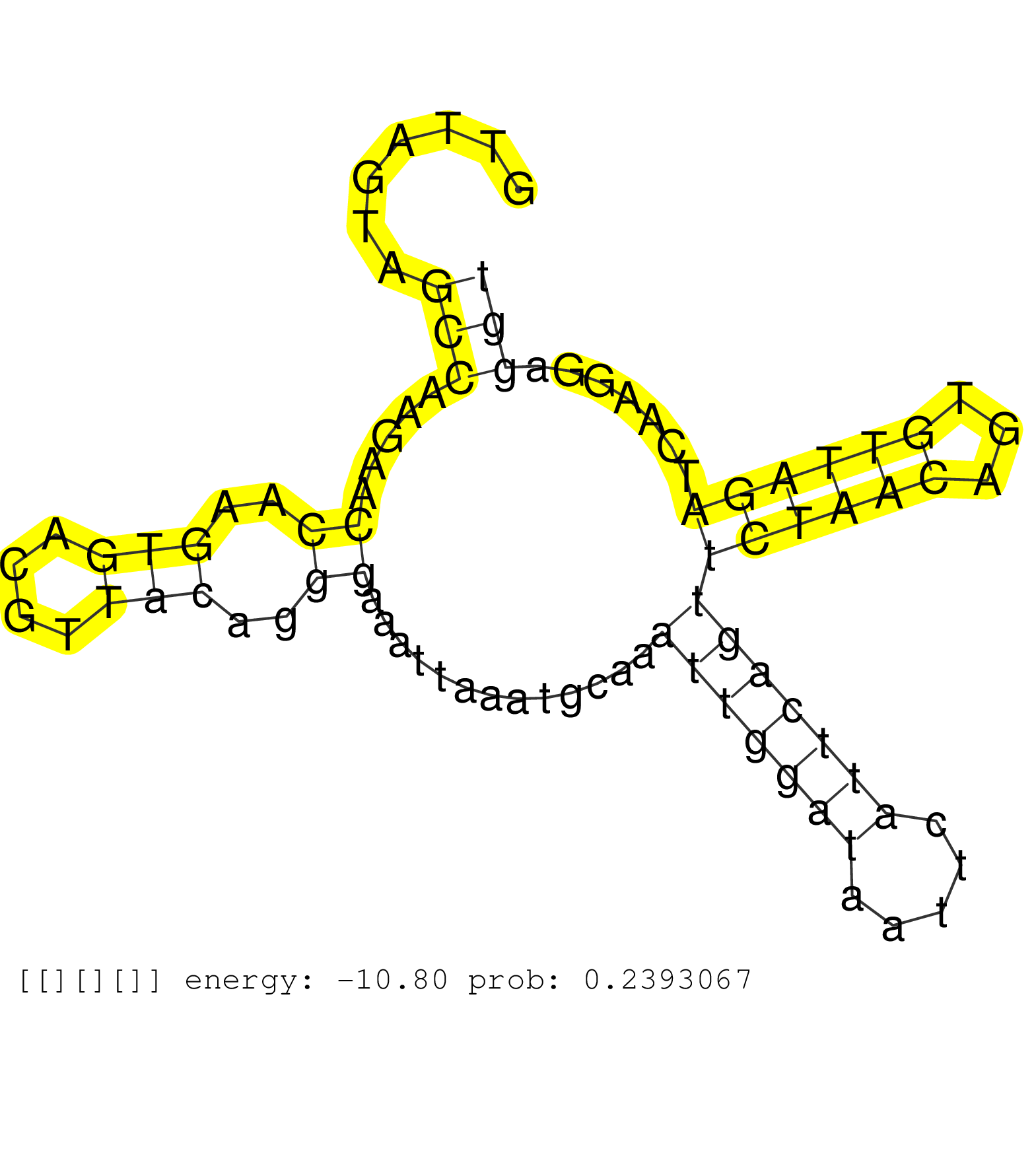

| Gene: Csnk1a1 | ID: uc008fce.1_intron_8_0_chr18_61740142_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(13) TESTES |
| CCAAACCCCCACAGGCAAGCAAACTGACAAAACCAAGAGTAACATGAAAGGTTAGTAGCCAAGAACCAAGTGACGTTACAGGGAAATTAAATGCAAATTGGATAATTCATTCAGTTCTAACAGTGTTAGATCAAGGAGGTGGTTTTAAAACATAAAATTTGGCTTCATATTTAAAAAAAAAAAAAAGACGTCCTTGGAAAATTTGACTACTAACTTTAAACCCAAATGTCCTTGTTCATATATATGTATA .........................................................(((.....((..(((....)))..)).............(((((((.....)))))))((((((...)))))).......))).............................................................................................................. ..................................................51.......................................................................................140............................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTTAGTAGCCAAGAACCAAGTGACGTT............................................................................................................................................................................. | 27 | 1 | 14.00 | 14.00 | 6.00 | 5.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ..................................................GTTAGTAGCCAAGAACCAAGTGACGT.............................................................................................................................................................................. | 26 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............AGGCAAGCAAACTGACAAAACCAAGAG................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............AGGCAAGCAAACTGACAAAACCAAGAGT.................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GTGTTAGATCAAGGAGGTG............................................................................................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................GTTAGATCAAGGAGGTG............................................................................................................. | 17 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTTAGTAGCCAAGAACCAAGTGACGgt............................................................................................................................................................................. | 27 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTTAGTAGCCAAGAACCAAGTGACGTTA............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTTAGTAGCCAAGAACCAAGTGACGTTAC........................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TAACAGTGTTAGATCAAGGAGGTGGTTT......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TAACAGTGTTAGATCAAGGAGGTGGTT.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............GCAAGCAAACTGACAAAACCAAGAGTAACA.............................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................TAGTAGCCAAGAACCAAGTGACGTTACA.......................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....CCCCCACAGGCAAGCAAACTGACAAAACC........................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................TGAAAGGTTAGTAGCCAAGAACCAAGTG.................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CTAACAGTGTTAGATCAAGG.................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................TAGTAGCCAAGAACCAAGTGACGTTAC........................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGTTAGATCAAGGtag............................................................................................................... | 16 | tag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....CCCCCACAGGCAAGCAAACTGACAAAACCA....................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TTAGTAGCCAAGAACCAAG.................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........CAGGCAAGCAAACTGACAAAACCAAGAG................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| CCAAACCCCCACAGGCAAGCAAACTGACAAAACCAAGAGTAACATGAAAGGTTAGTAGCCAAGAACCAAGTGACGTTACAGGGAAATTAAATGCAAATTGGATAATTCATTCAGTTCTAACAGTGTTAGATCAAGGAGGTGGTTTTAAAACATAAAATTTGGCTTCATATTTAAAAAAAAAAAAAAGACGTCCTTGGAAAATTTGACTACTAACTTTAAACCCAAATGTCCTTGTTCATATATATGTATA .........................................................(((.....((..(((....)))..)).............(((((((.....)))))))((((((...)))))).......))).............................................................................................................. ..................................................51.......................................................................................140............................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|