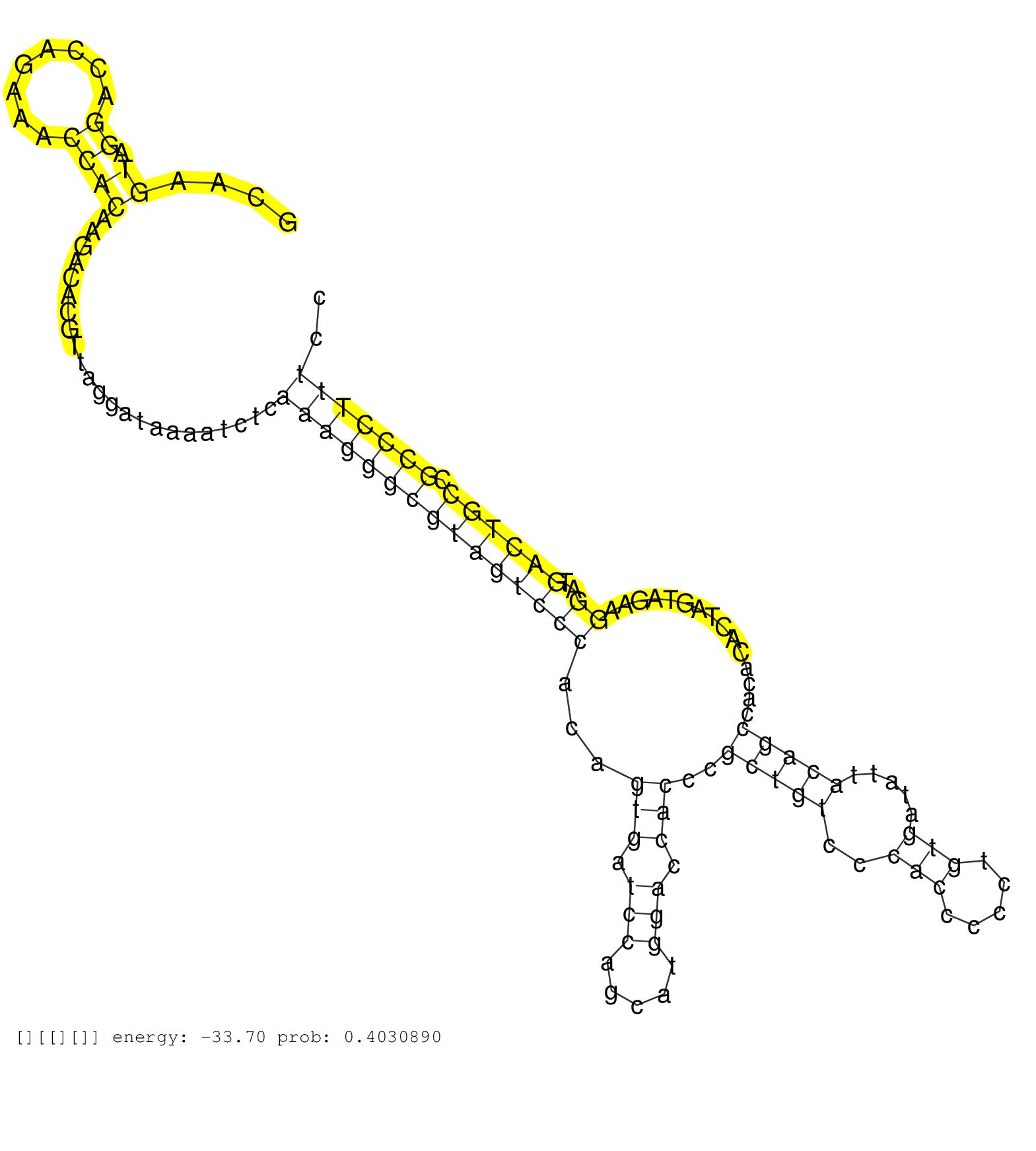
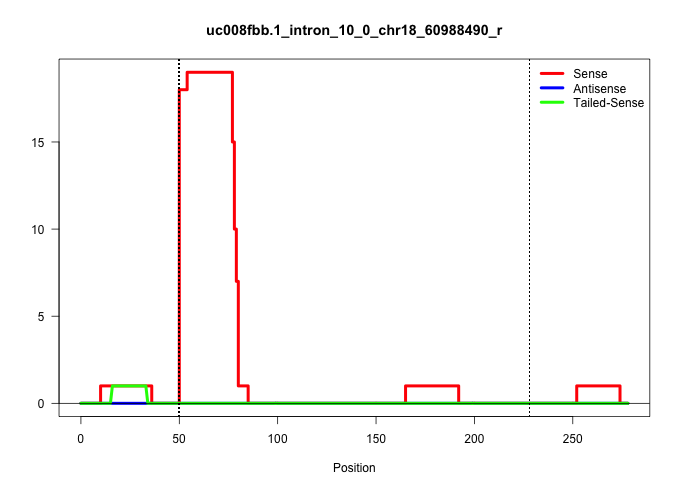
| Gene: Tcof1 | ID: uc008fbb.1_intron_10_0_chr18_60988490_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| AGACACCTCCAGCGTCTGCATCTGGAAAAGCTGTGGCTGCTCCAACCAAGGCAAGTAGGACCAGAAACCACAAGACACGTTAGGATAAAATCTCAAAGGGCGTAGTCCCACAGTGATCCAGCATGGACCACCCGCTGTCCCACCCCCTGTGATATTACAGCCACACACTAGTAGAAGGATGACTGCCGCCCTTTCCTCCTCCAATACCATTGCCACCTCTCGATTCAGGGAAAACCACCTGTTCCGAACAGCACCGTCTCTGCAAGGGGCCAGCGGTC ......................................................((.((........)))).......................(((((((((((((((...(((.(((.....))).)))..(((((..(((.....))).....)))))...............))..)))))).))))))).................................................................................... ..................................................51...............................................................................................................................................196................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GCAAGTAGGACCAGAAACCACAAGACACGT...................................................................................................................................................................................................... | 30 | 1 | 6.00 | 6.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GCAAGTAGGACCAGAAACCACAAGACAC........................................................................................................................................................................................................ | 28 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 |
| ..................................................GCAAGTAGGACCAGAAACCACAAGACA......................................................................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..................................................GCAAGTAGGACCAGAAACCACAAGACACG....................................................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................CACTAGTAGAAGGATGACTGCCGCCCT...................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTAGGACCAGAAACCACAAGACACGTTAGGA................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................ACCGTCTCTGCAAGGGGCCAGC.... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................TGCATCTGGAAAAGacgt.................................................................................................................................................................................................................................................... | 18 | acgt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........AGCGTCTGCATCTGGAAAAGCTGTGG.................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| AGACACCTCCAGCGTCTGCATCTGGAAAAGCTGTGGCTGCTCCAACCAAGGCAAGTAGGACCAGAAACCACAAGACACGTTAGGATAAAATCTCAAAGGGCGTAGTCCCACAGTGATCCAGCATGGACCACCCGCTGTCCCACCCCCTGTGATATTACAGCCACACACTAGTAGAAGGATGACTGCCGCCCTTTCCTCCTCCAATACCATTGCCACCTCTCGATTCAGGGAAAACCACCTGTTCCGAACAGCACCGTCTCTGCAAGGGGCCAGCGGTC ......................................................((.((........)))).......................(((((((((((((((...(((.(((.....))).)))..(((((..(((.....))).....)))))...............))..)))))).))))))).................................................................................... ..................................................51...............................................................................................................................................196................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................CCAGCATGGACCACttag................................................................................................................................................... | 18 | ttag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |