
| Gene: Ap3s1 | ID: uc008evv.1_intron_1_0_chr18_46914117_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
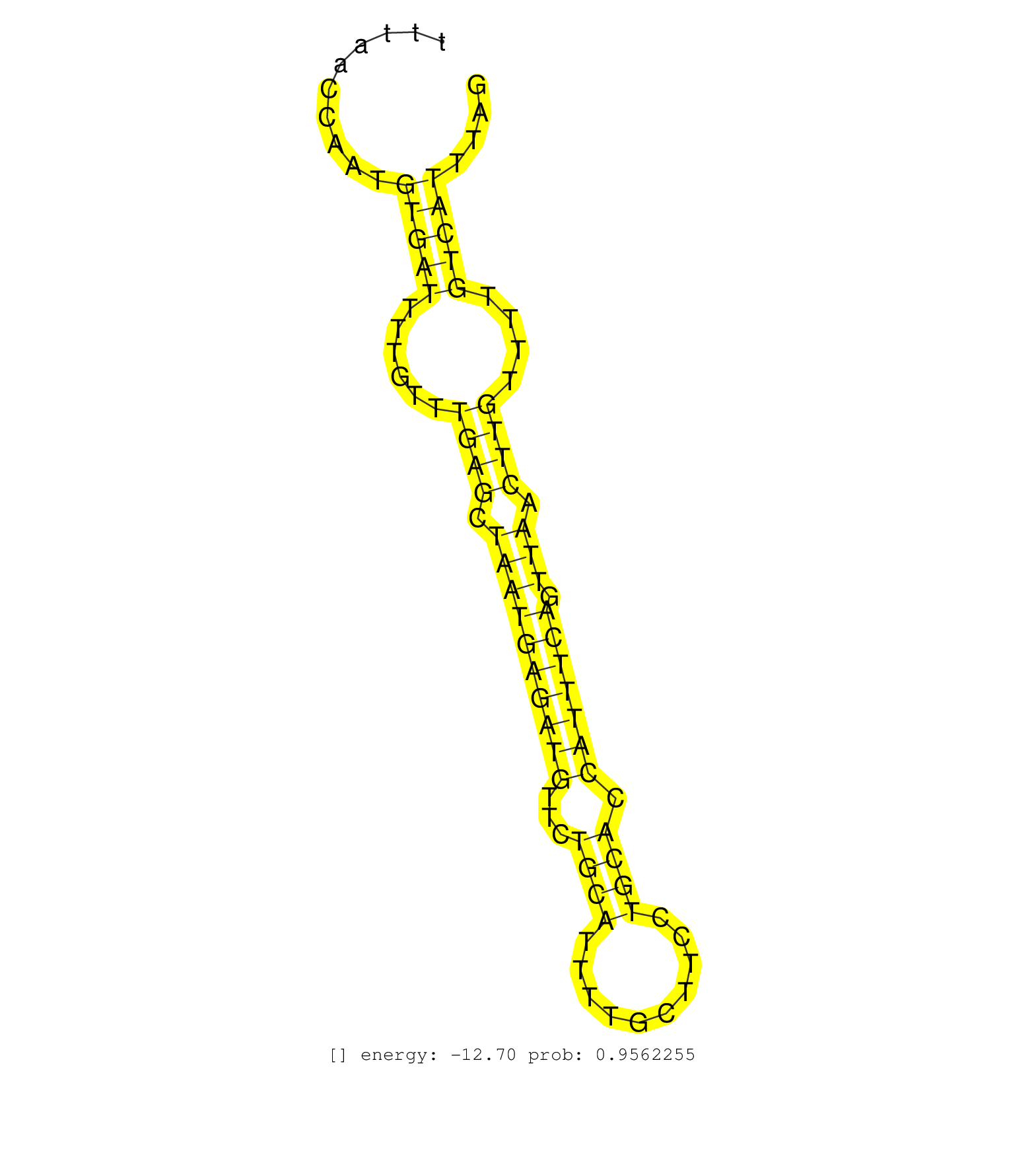
| CACTGGTTTCTTTTACTTGACATTTCTGAAGAAAATTGCTATCTTAATTTAGTTTCTGAAGTAAGTTTACTTGAATGCTATGCCATTGTTATATATTTTGTTTTATAAAAGTTTTTAACCAATGTGATTTTGTTTGAGCTAATGAGATGTTCTGCATTTTGCTTCCTGCACCATTTCAGTTAACTTGTTTTGTCATTTAGATTAATTGGAGGCTCTGACAACAAGCTCATTTACAGACATTATGCAACAC ...........................................................................................................................(((((......((((.((((((((((...((((..........)))).))))))).))).))))....)))))...................................................... .................................................................................................................114...................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................CCAATGTGATTTTGTTTGAGCTAATGAGATGTTCTGCATTTTGCTTCCTGCA................................................................................ | 52 | 1 | 45.00 | 45.00 | 45.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGACAACAAGCTCATTTACAGACATTA........ | 27 | 3 | 1.33 | 1.33 | - | - | 0.67 | - | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTAATTGGAGGCTCTGACAACAAGCTC...................... | 27 | 3 | 1.00 | 1.00 | - | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| ..........................................................................................................................................................................................................TAATTGGAGGCTCTGACAAaa........................... | 21 | aa | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TCAGTTAACTTGTTTTGTCATTTAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TCTGACAACAAGCTCATTTACAGACATT......... | 28 | 3 | 0.67 | 0.67 | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ATTAATTGGAGGCTCTGACAACAAGCTC...................... | 28 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 | - | - | - | - | - |
| ...........................................................................................................................................................................................................AATTGGAGGCTCTGACAACAAGCTC...................... | 25 | 3 | 0.67 | 0.67 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ..........................................................................................................................................................................................................TAATTGGAGGCTCTGACAACAAGCTCATT................... | 29 | 3 | 0.67 | 0.67 | - | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TTGGAGGCTCTGACAACAAGCTCATTTA................. | 28 | 3 | 0.67 | 0.67 | - | - | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGACAACAAGCTCATTTACAGACATTAT....... | 28 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAATTGGAGGCTCTGACAACAAGCTCA..................... | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................ATTGGAGGCTCTGACAACAAG......................... | 21 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| .....................................................................................................................................................................................................................TCTGACAACAAGCTCATTTACAGACAT.......... | 27 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAATTGGAGGCTCTGACAACAAGCTC...................... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TTGGAGGCTCTGACAACAAGCTCATTT.................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CTCTGACAACAAGCTCATTTACAGACAT.......... | 28 | 3 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TCTGACAACAAGCTCATTTACAGACA........... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CTCTGACAACAAGCTCATTTACAGACA........... | 27 | 3 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGGAGGCTCTGACAACAAGCTCATTTA................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ...........................................................................................................................................................................................................AATTGGAGGCTCTGACAACAAGCT....................... | 24 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGGAGGCTCTGACAACAAGCT....................... | 21 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAATTGGAGGCTCTGACAAC............................ | 20 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| CACTGGTTTCTTTTACTTGACATTTCTGAAGAAAATTGCTATCTTAATTTAGTTTCTGAAGTAAGTTTACTTGAATGCTATGCCATTGTTATATATTTTGTTTTATAAAAGTTTTTAACCAATGTGATTTTGTTTGAGCTAATGAGATGTTCTGCATTTTGCTTCCTGCACCATTTCAGTTAACTTGTTTTGTCATTTAGATTAATTGGAGGCTCTGACAACAAGCTCATTTACAGACATTATGCAACAC ...........................................................................................................................(((((......((((.((((((((((...((((..........)))).))))))).))).))))....)))))...................................................... .................................................................................................................114...................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................ATGTGATTTTGTTTcgca................................................................................................................... | 18 | cgca | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |