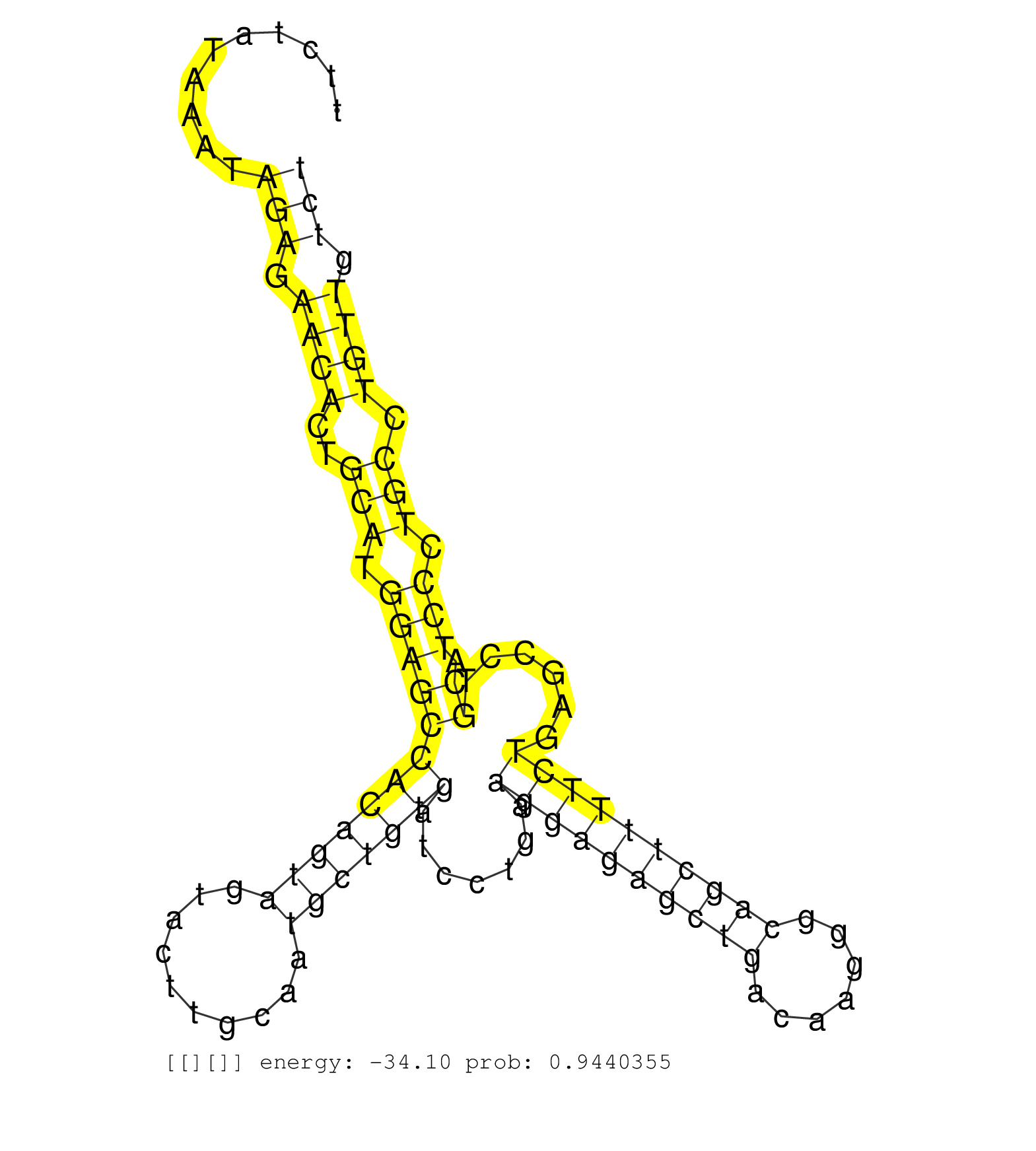
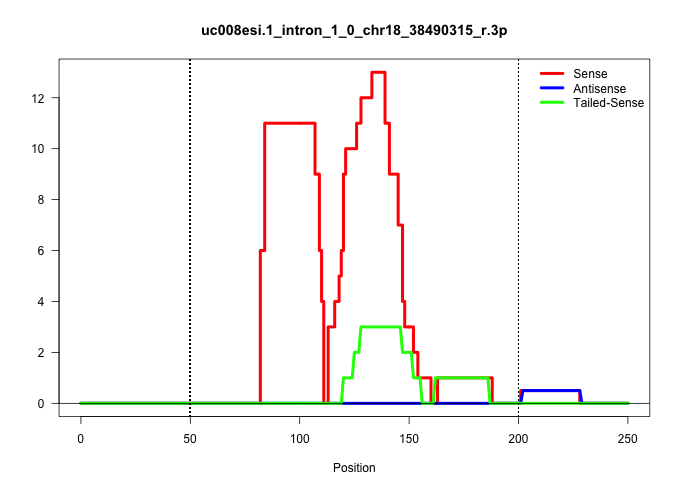
| Gene: Gnpda1 | ID: uc008esi.1_intron_1_0_chr18_38490315_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(16) TESTES |
| GTCTGTAGAGTGAGTTCTTAGGGTCACCTCAGCACCTCTGCTCCTGGTGTCCTAGCTCCAGTGGGGCCATGCCTTTGCTTTCTATAAATAGAGAACACTGCATGGAGCCACAGTAGTACTTGCAATGCTGTGATCCTGAAGGAGAGCTGACAAGGGCAGCTTTTCTGAGCCTGCATCCCTGCCTGTTGTCTGATCCCCAGGTGATGATCCTCATCACAGGCGCTCACAAGGCCTTTGCTCTGTACAAAGC .........................................................................................(((.((((..(((.((((((((((((..........))))))).......((((((((((.......))))))))))......)).))).))).)))).)))........................................................... ...............................................................................80.............................................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................TAAATAGAGAACACTGCATGGAGCCAC........................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................TATAAATAGAGAACACTGCATGGAGCC............................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................TGCAATGCTGTGATCCTGAAGGAGAGC....................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| ..................................................................................TATAAATAGAGAACACTGCATGGAG............................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................TAGTACTTGCAATGCTGTGATCCTGA............................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TAAATAGAGAACACTGCATGGAGCCA............................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TATAAATAGAGAACACTGCATGGAGCCAC........................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TTCTGAGCCTGCATCCCTGCCTGTa............................................................... | 25 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TCTGAGCCTGCATCCCTGCCTGTTG.............................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................TGCTGTGATCCTGAAGGAGAGCTGAtt.................................................................................................. | 27 | tt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCAATGCTGTGATCCTGAAGGAGAGCT...................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TCCTGAAGGAGAGCTGACAAGGGCAGC.......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................TAGTACTTGCAATGCTGTGATCCTGAAG............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TGTGATCCTGAAGGAGAGCTGACAAGac.............................................................................................. | 28 | ac | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................TACTTGCAATGCTGTGATCCTGAAG............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GCAATGCTGTGATCCTGAAGGAGAGC....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TTGCAATGCTGTGATCCTGAAGGAGA......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCAATGCTGTGATCCTGAAGGAGAG........................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................GCTGTGATCCTGAAGGAGAGCTGACA.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCAATGCTGTGATCCTGAAGGAGcgc....................................................................................................... | 27 | cgc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TGTGATCCTGAAGGAGAGCTGACAAG................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................CTTGCAATGCTGTGATCCTGAAGGAGA......................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGATGATCCTCATCACAGGCGCTCACA...................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| GTCTGTAGAGTGAGTTCTTAGGGTCACCTCAGCACCTCTGCTCCTGGTGTCCTAGCTCCAGTGGGGCCATGCCTTTGCTTTCTATAAATAGAGAACACTGCATGGAGCCACAGTAGTACTTGCAATGCTGTGATCCTGAAGGAGAGCTGACAAGGGCAGCTTTTCTGAGCCTGCATCCCTGCCTGTTGTCTGATCCCCAGGTGATGATCCTCATCACAGGCGCTCACAAGGCCTTTGCTCTGTACAAAGC .........................................................................................(((.((((..(((.((((((((((((..........))))))).......((((((((((.......))))))))))......)).))).))).)))).)))........................................................... ...............................................................................80.............................................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................GATGATCCTCATCACAGGCGCTCACAA..................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |