
| Gene: Diap1 | ID: uc008erj.1_intron_4_0_chr18_38014259_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(14) TESTES |
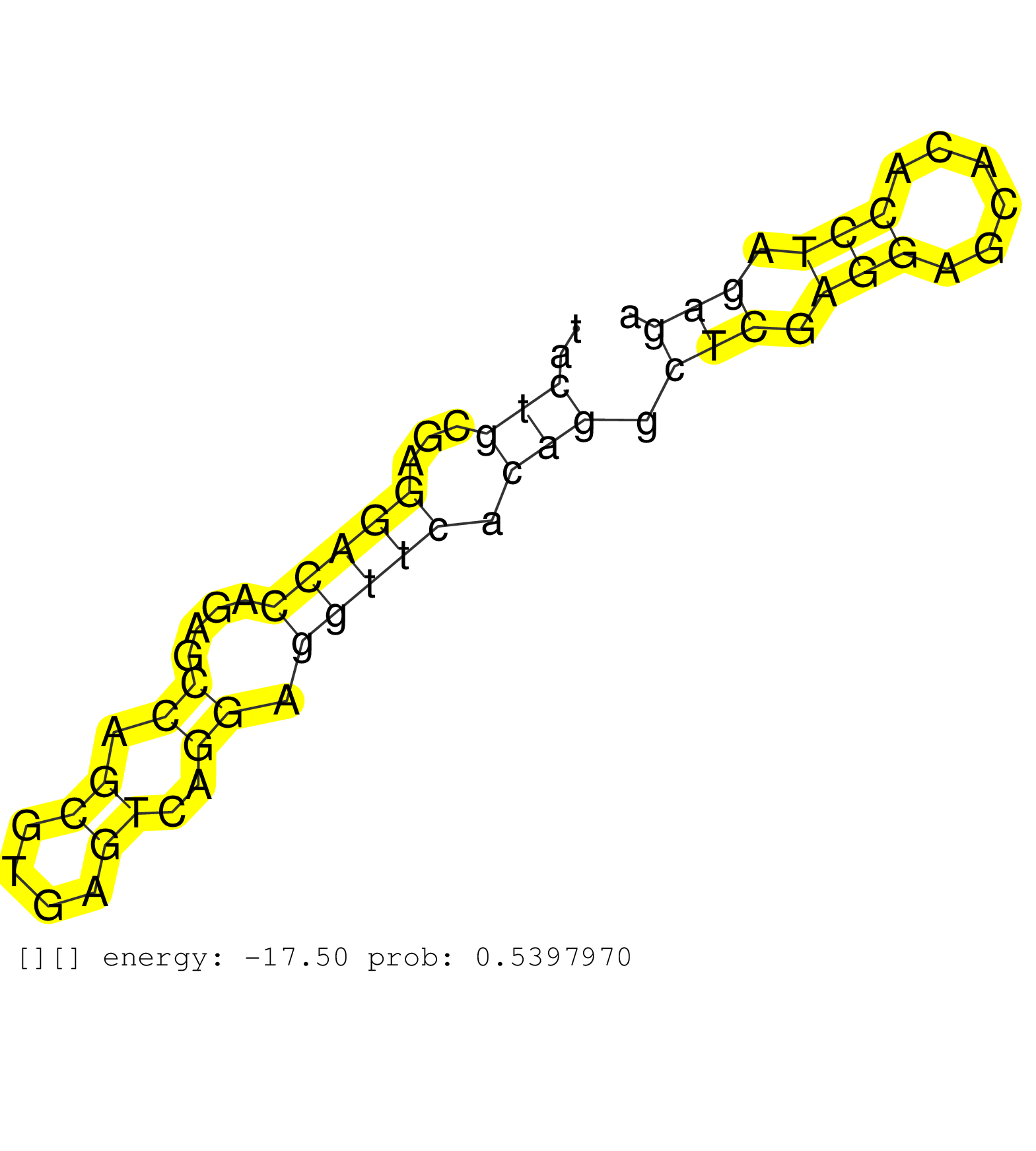
| TATACAGTGGGTAAAGCTTGCAAGTTCAGTTAGCAAGTACCCATGCTTTTACTGCGAGGACCAGAGCCAGCGTGAGTCAGGAGGTTCACAGGCTCGAGGAGCACACCTAGAGAAGAAAGGGAGGAGCGTAAGCAGGGCGGCTGGGGGGAGGCGCAGAGACTGAGCTGCTCAGTGCGCGCTTGTGTCTGTCTGGCTCCCAGTCTCTGCTGAGAACCTGCAGAAGAGCTTAGATCAGATGAAGAAGCAGATT ...................................................(((...(((((....((.((....))..)).))))).))).(((.(((......))).))).......................................................................................................................................... .................................................50.............................................................113....................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................TGAGAACCTGCAGAAGAGCTTAGATC................. | 26 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGAGAACCTGCAGAAGAGCTTAGATCA................ | 27 | 1 | 5.00 | 5.00 | 1.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTGTGTCTGTCTGGCTCCCAGaa................................................ | 23 | aa | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTGTGTCTGTCTGGCTCCCAGa................................................. | 22 | a | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGAGAACCTGCAGAAGAGCTTAGATCAGA.............. | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGCTGAGAACCTGCAGAAGAGCTTA..................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| .............................................................................................TCGAGGAGCACACagt............................................................................................................................................. | 16 | agt | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTGTGTCTGTCTGGCTCCCAGT................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GAAGAGCTTAGATCAGATGAAGAAGC..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGAGAACCTGCAGAAGAGCTTt..................... | 22 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGAGAACCTGCAGAAGAGCTTAGATCAG............... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGCTGAGAACCTGCAGAAGAGCTTAG.................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................CGAGGACCAGAGCCAGCGTGAGTCAGGA........................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................TGAGAACCTGCAGAAGAGCTTAGATCAGAT............. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................CTGCTGAGAACCTGCAGAA............................ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGCTGAGAACCTGCAGAAGAGCTTAGAT.................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTTGTGTCTGTCTGGCTCCCAGT................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................................AGAGCTTAGATCAGATGAAGAAG...... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TCGAGGAGCACACagtc............................................................................................................................................ | 17 | agtc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTGTGTCTGTCTGGCTCCCAGata............................................... | 24 | ata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................TGAGAACCTGCAGAAGAGCTTAGATCAGt.............. | 29 | t | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AACCTGCAGAAGAGCTTAGATCAGATG............ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| TATACAGTGGGTAAAGCTTGCAAGTTCAGTTAGCAAGTACCCATGCTTTTACTGCGAGGACCAGAGCCAGCGTGAGTCAGGAGGTTCACAGGCTCGAGGAGCACACCTAGAGAAGAAAGGGAGGAGCGTAAGCAGGGCGGCTGGGGGGAGGCGCAGAGACTGAGCTGCTCAGTGCGCGCTTGTGTCTGTCTGGCTCCCAGTCTCTGCTGAGAACCTGCAGAAGAGCTTAGATCAGATGAAGAAGCAGATT ...................................................(((...(((((....((.((....))..)).))))).))).(((.(((......))).))).......................................................................................................................................... .................................................50.............................................................113....................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................ACCAGAGCCAGCGTGga................................................................................................................................................................................ | 17 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |