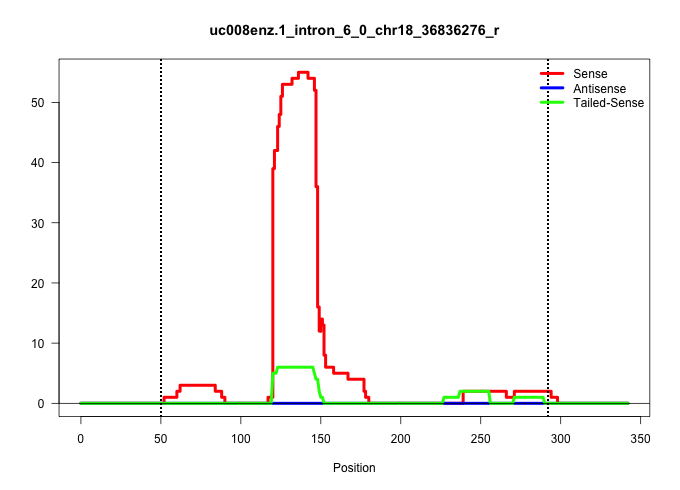
| Gene: Apbb3 | ID: uc008enz.1_intron_6_0_chr18_36836276_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(5) OTHER.mut |
(7) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(24) TESTES |
| AGCCTCTGGTTCACATCCGTGTGTGGGGTGTGGGCAGCTCCAAGGGCCGGTGAGGACCTTTGCATTTCCCCAAGCGAGAGTCGCCTCCCCTTCCCCCCTGCAGGCTGTGATCCCCACTGATGCTGATGCTGAACCCTTGGACACGGATCCTCTGATGTGGACCCAGCACGCCTTCCCTGCCTGCTTCATCCCTGGTTGGGGTGGGTGGGCCCATCTCAGCAGGGACAGATAGGGGAGAGTCTGAAAGACTGGGCTGATTTTCTCCTGTGCTCCCACATGCTTCTGGAACAGTGACAGGTATGTGATAACCAGGTACTCCAAGCCCTATCCATCCCTGACTTC ....................................................................................................................(((.(((((...(((((.....((((((((.(((........)))..((((....((((((((.(((.....(((((.(((((((((((....))))))))))))))))))).))))))))((((.....))))..))))........))))).))).......))).))..))))).)))............................................. ...................................................................................................................116.......................................................................................................................................................................................302...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................TGCTGATGCTGAACCCTTGGACACGGAT.................................................................................................................................................................................................. | 28 | 1 | 18.00 | 18.00 | 3.00 | 5.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCTGATGCTGAACCCTTGGACACGGA................................................................................................................................................................................................... | 27 | 1 | 15.00 | 15.00 | 5.00 | - | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCTGATGCTGAACCCTTGGACACGGATC................................................................................................................................................................................................. | 29 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CACGGATCCTCTGATGTGG...................................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCTGATGCTGAACCCTTGGACACGGttt................................................................................................................................................................................................. | 29 | ttt | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GATGCTGAACCCTTGGACACGGATCCTC.............................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TCTGATGTGGACCCAGCACGCCTTCCC..................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCTGATGCTGAACCCTTGGACACGG.................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGATGCTGAACCCTTGGACACGGATCCTC.............................................................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TTGGACACGGATCCTCTGATGTGGACCCAGC............................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGATGCTGAACCCTTGGACACGGATCCTCT............................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CATTTCCCCAAGCGAGAGTCGCCTCCCC............................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................TGCTGAACCCTTGGACACGGATCCTCT............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................CCCACATGCTTCTGGAACAGTGACAGG............................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................ATGCTGAACCCTTGGACACGGATCC................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGACCTTTGCATTTCCCCAAGCGAGAGTCGC.................................................................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................TCTGAAAGACTGGGCTGATTTTCTCC............................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................ATGCTGAACCCTTGGACACGGATCCT............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................TCTGAAAGACTGGGCTGATTTTCTCCT............................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................TGCTGAACCCTTGGACACGGATCCTC.............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GCTGATGCTGAACCCTTGGACACGGATC................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................ATGCTGAACCCTTGGACACGGATCCTC.............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................TGTGCTCCCACATGCTTCTGGAACAGTGA................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................TGCATTTCCCCAAGCGAGAGTCGCCTCC.............................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................CCCACATGCTTCTGGAttt.................................................... | 19 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................GCTGATGCTGAACCCTTGGACACGGAT.................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TGATGTGGACCCAGCACGCCTTCCCTGC.................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................GCTGATGCTGAACCCTTGGACACGGA................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGATGCTGATGCTGAACCCTTGGAC........................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGATGCTGAACCCTTGGACACGGAT.................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCTGATGCTGAACCCTTGGACACGGc................................................................................................................................................................................................... | 27 | c | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCTGATGCTGAACCCTTGGACACGGATCt................................................................................................................................................................................................ | 30 | t | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACCCTTGGACACGGATCCTCTGATGT........................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................................................................GATAGGGGAGAGTCTGAAAGACTGGGCTt...................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TCTGATGTGGACCCAGCACGCCTTCCCT.................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................AGTCTGAAAGACTGGatgt...................................................................................... | 19 | atgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGATGCTGAACCCTTGGACACGGATCCaa.............................................................................................................................................................................................. | 29 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCTGATGCTGAACCCTTGGACACGa.................................................................................................................................................................................................... | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| AGCCTCTGGTTCACATCCGTGTGTGGGGTGTGGGCAGCTCCAAGGGCCGGTGAGGACCTTTGCATTTCCCCAAGCGAGAGTCGCCTCCCCTTCCCCCCTGCAGGCTGTGATCCCCACTGATGCTGATGCTGAACCCTTGGACACGGATCCTCTGATGTGGACCCAGCACGCCTTCCCTGCCTGCTTCATCCCTGGTTGGGGTGGGTGGGCCCATCTCAGCAGGGACAGATAGGGGAGAGTCTGAAAGACTGGGCTGATTTTCTCCTGTGCTCCCACATGCTTCTGGAACAGTGACAGGTATGTGATAACCAGGTACTCCAAGCCCTATCCATCCCTGACTTC ....................................................................................................................(((.(((((...(((((.....((((((((.(((........)))..((((....((((((((.(((.....(((((.(((((((((((....))))))))))))))))))).))))))))((((.....))))..))))........))))).))).......))).))..))))).)))............................................. ...................................................................................................................116.......................................................................................................................................................................................302...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|