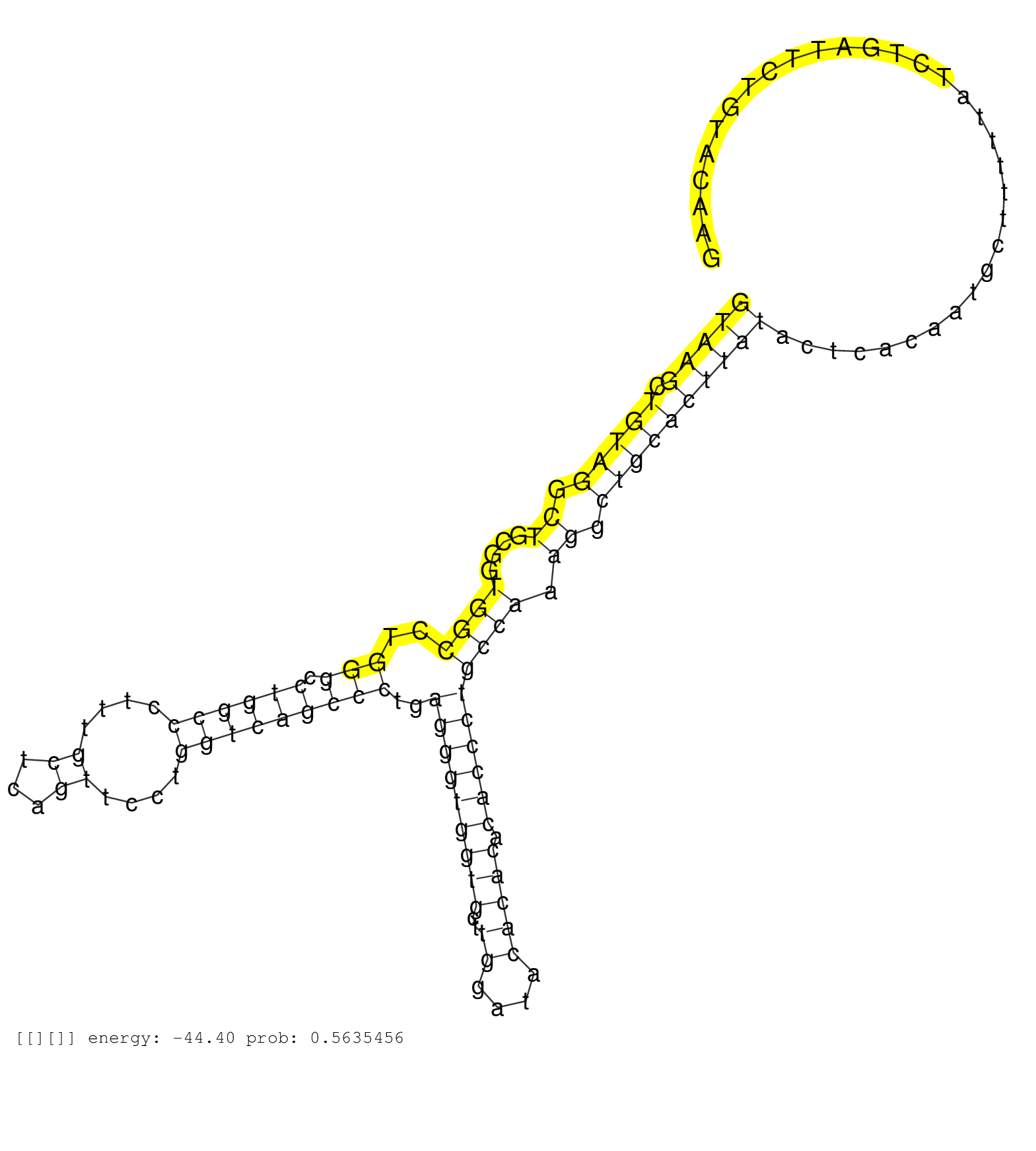

| Gene: Kif20a | ID: uc008ela.1_intron_10_0_chr18_34788939_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(21) TESTES |
| CACACCCTGGGCCGCTGTATTGCTGCCCTGCGACAGAATCAGCAGAACCGGTAAGCTGTAGGCTGCGGTGGCCTGGGCCTGGCCCTTTGCTCAGTTCCTGGTCAGCCCTGAGGGTGGTGCTTGGATACACACACACCCTGCCAAAGGCTGCACTTATACTCACAATGCTTTTTATCTGATTCTGTACAAGGTCAAAGCAGAACCTGATTCCTTTCCGTGACAGCAAGTTGACTCGTGTGT ..................................................(((((.(((((.((....((((..(((.((((((....((...))....)))))))))..(((((((((..((....))))).)))))))))).)).))))))))))................................................................................... ..................................................51.........................................................................................................................................190................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGCTGTAGGCTGCGGTGGCCTGG.................................................................................................................................................................... | 26 | 1 | 13.00 | 13.00 | 2.00 | 4.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTAAGCTGTAGGCTGCGGTGGCCTGGG................................................................................................................................................................... | 27 | 1 | 8.00 | 8.00 | 1.00 | 1.00 | - | - | 1.00 | - | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCTGTAGGCTGCGGTGGCCTGa.................................................................................................................................................................... | 26 | a | 3.00 | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCTGTAGGCTGCGGTGGCCTGGGC.................................................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGCCCTGCGACAGAATCAGCAGAACCGGT............................................................................................................................................................................................ | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TCCGTGACAGCAAGTTGACTCGTGTG. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGAACCTGATTCCTTTCCGTGACAGC................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGCCCTGCGACAGAATCAGCAGAACCGGTc........................................................................................................................................................................................... | 30 | c | 2.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCTGTAGGCTGCGGTGGCC....................................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................TGCGACAGAATCAGCAGAACCGGTca.......................................................................................................................................................................................... | 26 | ca | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAAGCTGTAGGCTGCGGTGGCCTGGt................................................................................................................................................................... | 27 | t | 1.00 | 13.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TCTGATTCTGTACAAG.................................................. | 16 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CAAAGCAGAACCTGATTCCTTTCCGTG..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TTCCTTTCCGTGACAGCAAGTTGACTCGTGTG. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................GCCCTGCGACAGAATCAGCA.................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGATTCCTTTCCGTGACAGCAAGT............ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................ACCGGTAAGCTGTAGGCTGCGGTGGC........................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGCGACAGAATCAGCAGAACCGGTcaa......................................................................................................................................................................................... | 27 | caa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCTGTAGGCTGCGGTGGCCTG..................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CAAAGCAGAACCTGATTCCTTTCCGT...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CAAAGCAGAACCTGATTCCTT........................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................CAGAATCAGCAGAACCG.............................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TCCTTTCCGTGACAGCAAG............. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................ACCGGTAAGCTGTAGGCTGCGG............................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TTCCGTGACAGCAAGTTGACTCGTGTG. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CAGCAAGTTGACTCGTGT.. | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTGTACAAGGTCAAAGCAGAACCTG.................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................AGGCTGCGGTGGCCTGGGCCTGG.............................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTAAGCTGTAGGCTGCGGTGGCCT...................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................AAAGCAGAACCTGATTCCTTTCCGTGAC................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................ACAGAATCAGCAGAACCGG............................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACACCCTGGGCCGCTGTATTGCTGCCCTGCGACAGAATCAGCAGAACCGGTAAGCTGTAGGCTGCGGTGGCCTGGGCCTGGCCCTTTGCTCAGTTCCTGGTCAGCCCTGAGGGTGGTGCTTGGATACACACACACCCTGCCAAAGGCTGCACTTATACTCACAATGCTTTTTATCTGATTCTGTACAAGGTCAAAGCAGAACCTGATTCCTTTCCGTGACAGCAAGTTGACTCGTGTGT ..................................................(((((.(((((.((....((((..(((.((((((....((...))....)))))))))..(((((((((..((....))))).)))))))))).)).))))))))))................................................................................... ..................................................51.........................................................................................................................................190................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................AACCTGATTCCTTTCCGTGACAGCA............... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................................................AACCTGATTCCTTTCCGTGACAGCAaa............... | 27 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CACACCCTGCCAAAGGCTGCACTTA.................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |