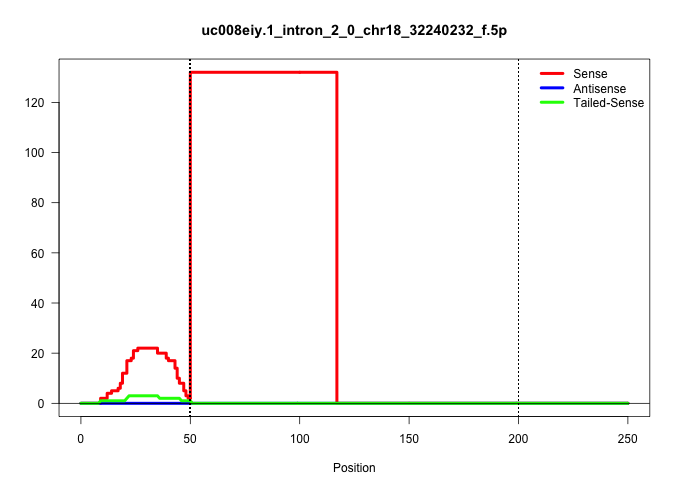
| Gene: Iws1 | ID: uc008eiy.1_intron_2_0_chr18_32240232_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) OVARY |
(4) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| GCGGAAGGCTGCTGTGCTTTCTGATAGTGAGGATGACGCAGGCAACGCATGTAAGCATTCCTATTGGACTGTCGCTTTCCTTATTGACTCTAGCTACAGCTAGAATACTGGGTCCTCTTCTGGTATGTTTTATTAGCATATATTGATTGTAATGTTAGTTTCTTCATGGCTATTTCACATAAGTACATAATGTATTTTAATCATACTTATCTCATTATCCTGTCTTGTCCAACTCCCTCTCATACTCAGT ........................................................................................................((((..((......))..))))......((((.((((((((..(((((((((((((.......)))))....))))...)))))))))))).)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGCATTCCTATTGGACTGTCGCTTTCCTTATTGACTCTAGCTACAGCTA.................................................................................................................................................... | 52 | 1 | 135.00 | 135.00 | 135.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TTCTGATAGTGAGGATGACGCAGGCAACGCA......................................................................................................................................................................................................... | 31 | 1 | 5.00 | 5.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - |
| ...................TCTGATAGTGAGGATGACGCAGGCA.............................................................................................................................................................................................................. | 25 | 1 | 5.00 | 5.00 | - | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ...............................GATGACGCAGGCAACGCA......................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - |
| .....................TGATAGTGAGGATGACGCAGGCAACGC.......................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................TAGTGAGGATGACGCAGGCAACGCAT........................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........TGCTGTGCTTTCTGATAGTGAGGATG....................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............TGTGCTTTCTGATAGTGAGGATGACGC................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .....................TGATAGTGAGGATGACGCAGGCAACG........................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TGATAGTGAGGATGACGCAGGCAACGCA......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................TTCTGATAGTGAGGATGACGCAGGC............................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................TAGTGAGGATGACGCAGGC............................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........CTGTGCTTTCTGATAGTGAGGATG....................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................GTGAGGATGACGCAGGCAACG........................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................TTTCTGATAGTGAGGATGACGCAGGC............................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................GATAGTGAGGATGACGCAGGCAACGCATc....................................................................................................................................................................................................... | 29 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............TGCTTTCTGATAGTGAGGATGACGCA.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AGCATATATTGATTGTAATGTTAG............................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................ATAGTGAGGATGACGCAGGCA.............................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................TTCTGATAGTGAGGATGACGCAGGCAA............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................TGATAGTGAGGATGACGCAGGCAAa............................................................................................................................................................................................................ | 25 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................TCTGATAGTGAGGATGACGCAGGCAA............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........TGCTGTGCTTTCTGATAGTGAGGATGt...................................................................................................................................................................................................................... | 27 | t | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| GCGGAAGGCTGCTGTGCTTTCTGATAGTGAGGATGACGCAGGCAACGCATGTAAGCATTCCTATTGGACTGTCGCTTTCCTTATTGACTCTAGCTACAGCTAGAATACTGGGTCCTCTTCTGGTATGTTTTATTAGCATATATTGATTGTAATGTTAGTTTCTTCATGGCTATTTCACATAAGTACATAATGTATTTTAATCATACTTATCTCATTATCCTGTCTTGTCCAACTCCCTCTCATACTCAGT ........................................................................................................((((..((......))..))))......((((.((((((((..(((((((((((((.......)))))....))))...)))))))))))).)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|