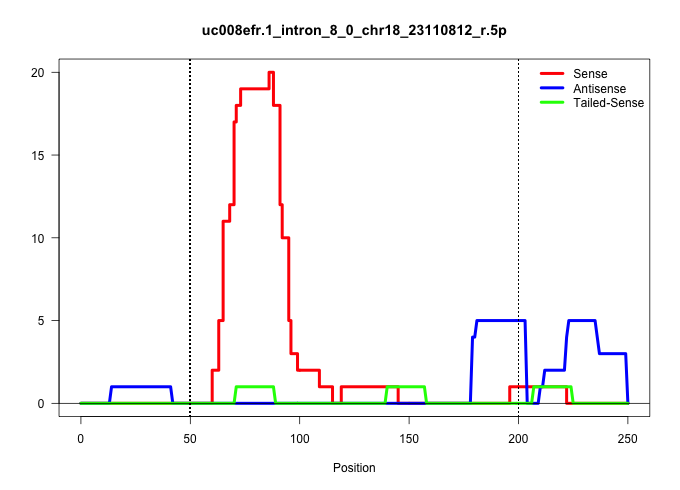
| Gene: Nol4 | ID: uc008efr.1_intron_8_0_chr18_23110812_r.5p | SPECIES: mm9 |
 |
|
(6) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| GGGGAGGCGGCGGCGCCAAGCAAGTGCTCTTCGTGCGGGTCAAGACCACGGTGAGCCGGTTTCTCTGTTATTATTTGTCTGTTGGAGCAGCCCGGAAGGGAGGGCGGAGAGAGGGGAAGGCAAAGAGATGCTGGAGACAGTTGGCTCCTAGCTCCCCCCCCCCCAGCCCTCCTCAACCCCCTCCCCAAGTCCTCCCTAGACGCAGTGGCCGGGCGTGATTTCCCTTCTAGAAAGTCTTTGTGGCTCTTTA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................TGTTATTATTTGTCTGTTGGAGCAGC............................................................................................................................................................... | 26 | 1 | 6.00 | 6.00 | 3.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................TTATTTGTCTGTTGGAGCAGCCCGG........................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................TTCTCTGTTATTATTTGTCTGTTGGAGC.................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TTATTTGTCTGTTGGAGCAGCCCGGA.......................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TCTGTTATTATTTGTCTGTTGGAGCAGCC.............................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................GCCGGGCGTGATTTCCtc......................... | 18 | tc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TATTTGTCTGTTGGggga................................................................................................................................................................. | 18 | ggga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................................................TAGACGCAGTGGCCGGGCGTGATTTC............................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TATTTGTCTGTTGGAGCAGCCCGG........................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TATTATTTGTCTGTTGGAGCAGCCCGG........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................GCAAAGAGATGCTGGAGACAGTTGGC......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................GCAGCCCGGAAGGGAGGGCGGAG............................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................CCGGAAGGGAGGGCGGAGAGAGGG....................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TCTGTTATTATTTGTCTGTTGGAGCAGC............................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TTGGCTCCTAGCTCagcc............................................................................................ | 18 | agcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................TTTGTCTGTTGGAGCAGCCCGGAAGG....................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| GGGGAGGCGGCGGCGCCAAGCAAGTGCTCTTCGTGCGGGTCAAGACCACGGTGAGCCGGTTTCTCTGTTATTATTTGTCTGTTGGAGCAGCCCGGAAGGGAGGGCGGAGAGAGGGGAAGGCAAAGAGATGCTGGAGACAGTTGGCTCCTAGCTCCCCCCCCCCCAGCCCTCCTCAACCCCCTCCCCAAGTCCTCCCTAGACGCAGTGGCCGGGCGTGATTTCCCTTCTAGAAAGTCTTTGTGGCTCTTTA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................CCTCCCCAAGTCCTCCCTAGACGCA.............................................. | 25 | 1 | 4.00 | 4.00 | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................CCTTCTAGAAAGTCTTTGTGGCTCTTTA | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..............GCCAAGCAAGTGCTCTTCGTGCGGGTCA................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CTTCTAGAAAGTCTTTGTGGCTCTTTA | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................GGGCGTGATTTCCCTTCTAGAAAGTC.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................GCGTGATTTCCCTTCTAGAAAGTCT............. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TCCCCAAGTCCTCCCTAGACGCA.............................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |