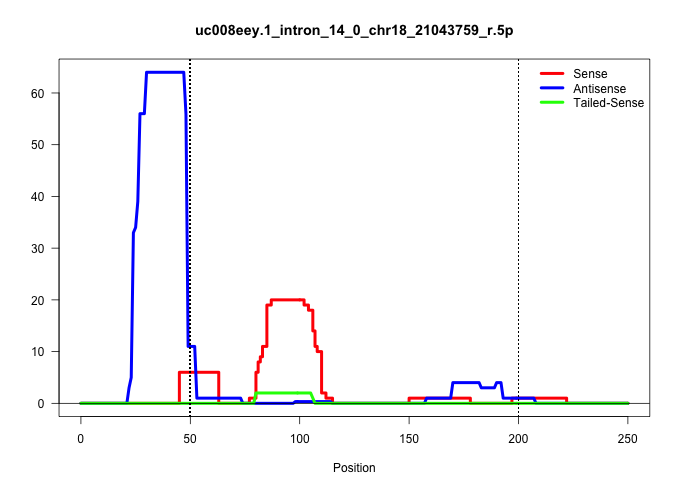
| Gene: D030074E01Rik | ID: uc008eey.1_intron_14_0_chr18_21043759_r.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| CCTCAGCTTCTCCGAGCTGCTCAAGCCCTTCTCTCGCCTCACTTCCGAAGGTACGTGGTGGCCTTCCTCGTTCACCAGGCTGAAACCGAGCTGTGGTCGAGAGAAGGAAGTTGGGAGCGAGCGGAGGACAGAGGGGCGGCGGCCTGGGGCTGCAGCCGGACGAGGCGCGGCGAGCGTCTGTCAAGACAGCGCGCTCTTGTCCGCCGGGCTGTCTAGGCGCTTGCTTCCTTGGACAGACCCTGGTCATTAA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................CCGAGCTGTGGTCGAGAGAAGGAAG............................................................................................................................................ | 25 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................CGAAGGTACGTGGTGGCC........................................................................................................................................................................................... | 18 | 1 | 6.00 | 6.00 | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAAACCGAGCTGTGGTCGAGAGAAG................................................................................................................................................ | 26 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - |
| .................................................................................GAAACCGAGCTGTGGTCGAGAGAAGG............................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................AACCGAGCTGTGGTCGAGAGAAGGA.............................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TGCAGCCGGACGAGGCGCGGCGAGCGTC........................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................AACCGAGCTGTGGTCGAGAGAAGG............................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GAGCTGTGGTCGAGAGAAGGAAGTT.......................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................TGAAACCGAGCTGTGGTCGAGAGAcgg............................................................................................................................................... | 27 | cgg | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TGTCCGCCGGGCTGTCTAGGCGCTT............................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAAACCGAGCTGTGGTCGAGAGA.................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................GGCTGAAACCGAGCTGTGGTCGAGA.................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................AAACCGAGCTGTGGTCGAGAGAAGGAAGTTGGG....................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAAACCGAGCTGTGGTCGAGAGAAa................................................................................................................................................ | 26 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCTCAGCTTCTCCGAGCTGCTCAAGCCCTTCTCTCGCCTCACTTCCGAAGGTACGTGGTGGCCTTCCTCGTTCACCAGGCTGAAACCGAGCTGTGGTCGAGAGAAGGAAGTTGGGAGCGAGCGGAGGACAGAGGGGCGGCGGCCTGGGGCTGCAGCCGGACGAGGCGCGGCGAGCGTCTGTCAAGACAGCGCGCTCTTGTCCGCCGGGCTGTCTAGGCGCTTGCTTCCTTGGACAGACCCTGGTCATTAA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................GCCCTTCTCTCGCCTCACTTCCGAA......................................................................................................................................................................................................... | 25 | 1 | 27.00 | 27.00 | 14.00 | 8.00 | 2.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................CTTCTCTCGCCTCACTTCCGAA......................................................................................................................................................................................................... | 22 | 1 | 17.00 | 17.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CTCTCGCCTCACTTCCGAAGGTA..................................................................................................................................................................................................... | 23 | 1 | 8.00 | 8.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................CCTTCTCTCGCCTCACTTCCGA.......................................................................................................................................................................................................... | 22 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AAGCCCTTCTCTCGCCTCACTTCCGA.......................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CGAGCGTCTGTCAAGACAGCGCG......................................................... | 23 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GCCCTTCTCTCGCCTCACTTCCGAAGGTA..................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................CTCTCGCCTCACTTCCGAAGGTAcag..................................................................................................................................................................................................... | 26 | cag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................CTTCCGAAGGTACGaag................................................................................................................................................................................................... | 17 | aag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................AGCCCTTCTCTCGCCTCACTTCCGA.......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GACGAGGCGCGGCGAGCGTCTGTCA................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................GCGCTCTTGTCCGCCGGG.......................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................AGCCCTTCTCTCGCCTCACTTCCGAA......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGGTACGTGGTGGCCTTCCTCGTTCA................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................CCCTTCTCTCGCCTCACTTCCGAAGGTA..................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................GCCGGACGAGGCGgcct................................................................................... | 17 | gcct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TCCGCCGGGCTGTgtcc...................................... | 17 | gtcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................GAGAGAAGGAAGTTGGG....................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |