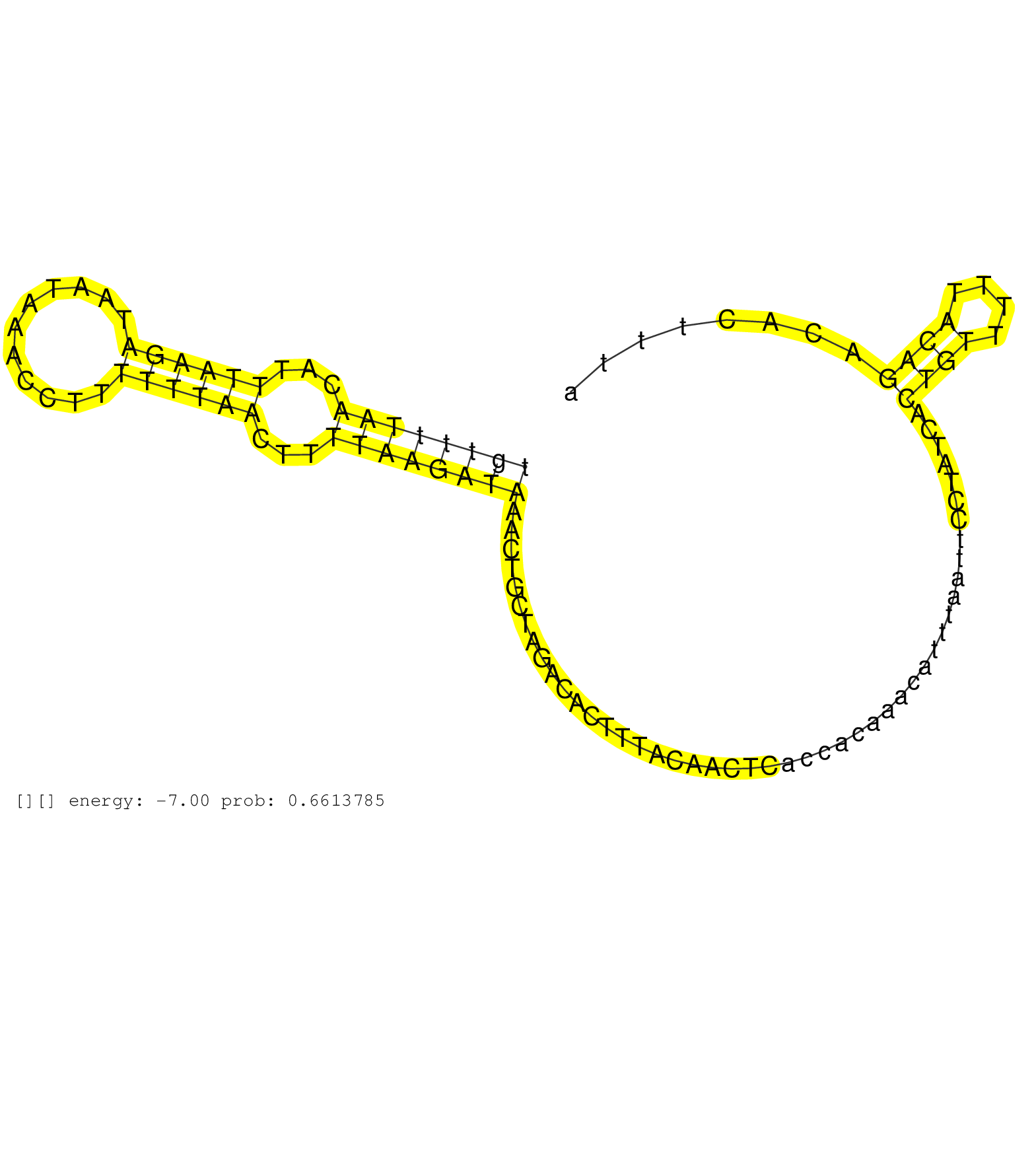
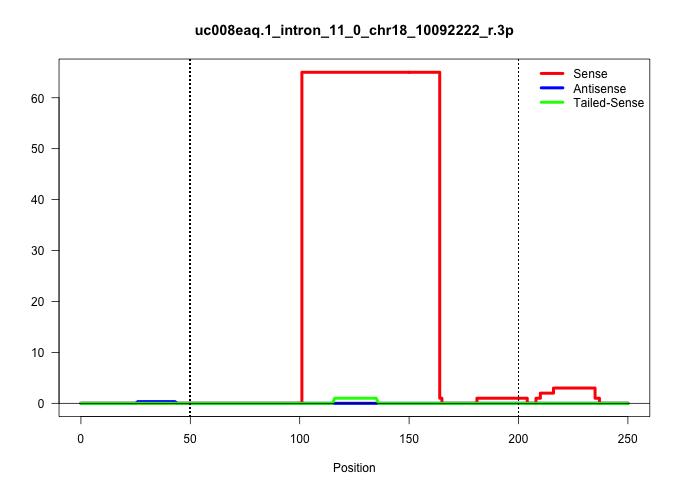
| Gene: Rock1 | ID: uc008eaq.1_intron_11_0_chr18_10092222_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(8) TESTES |
| AGCTGCCATGTGGGTGCTGGGAAGTGAACCTGGCTCTCAGGAAGAGCAGCCAGTGCTCTTAAACTGGTAAGCTATCTCTCTACTCCTGGTTTAACTTGTTTTAACATTTAAGATAATAAACCTTTTTTAACTTTTAAGATAAACTGCTAGACACTTTACAACTCACCACAAACATTTAATTCCTATCACTGTTTTTACAGACACTTTATAAAACCCAAGTAAAGGAGCTTAAAGAAGAAATTGAAGAAAA ................................................................................................((((((((...((((((...........))))))...))))))))...............................................((((....)))).................................................. ................................................................................................97.............................................................................................................208........................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................TAACATTTAAGATAATAAACCTTTTTTAACTTTTAAGATAAACTGCTAGACA................................................................................................. | 52 | 1 | 65.00 | 65.00 | 65.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AAGTAAAGGAGCTTAAAGA............... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................TAAACCTTTTTTAACTcaga.................................................................................................................. | 20 | caga | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AAACCCAAGTAAAGGAGCTTAAAGA............... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TAAAACCCAAGTAAAGGAGCTTAAAGAAG............. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................................CCTATCACTGTTTTTACAGACAC.............................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| AGCTGCCATGTGGGTGCTGGGAAGTGAACCTGGCTCTCAGGAAGAGCAGCCAGTGCTCTTAAACTGGTAAGCTATCTCTCTACTCCTGGTTTAACTTGTTTTAACATTTAAGATAATAAACCTTTTTTAACTTTTAAGATAAACTGCTAGACACTTTACAACTCACCACAAACATTTAATTCCTATCACTGTTTTTACAGACACTTTATAAAACCCAAGTAAAGGAGCTTAAAGAAGAAATTGAAGAAAA ................................................................................................((((((((...((((((...........))))))...))))))))...............................................((((....)))).................................................. ................................................................................................97.............................................................................................................208........................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................AGGAAGAGCAGCCAGTGCTtt................................................................................................................................................................................................. | 21 | tt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................CATTTAATTCCTAgagc................................................................. | 17 | gagc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - |
| ..........................AACCTGGCTCTCAGGAAG.............................................................................................................................................................................................................. | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | 0.33 |