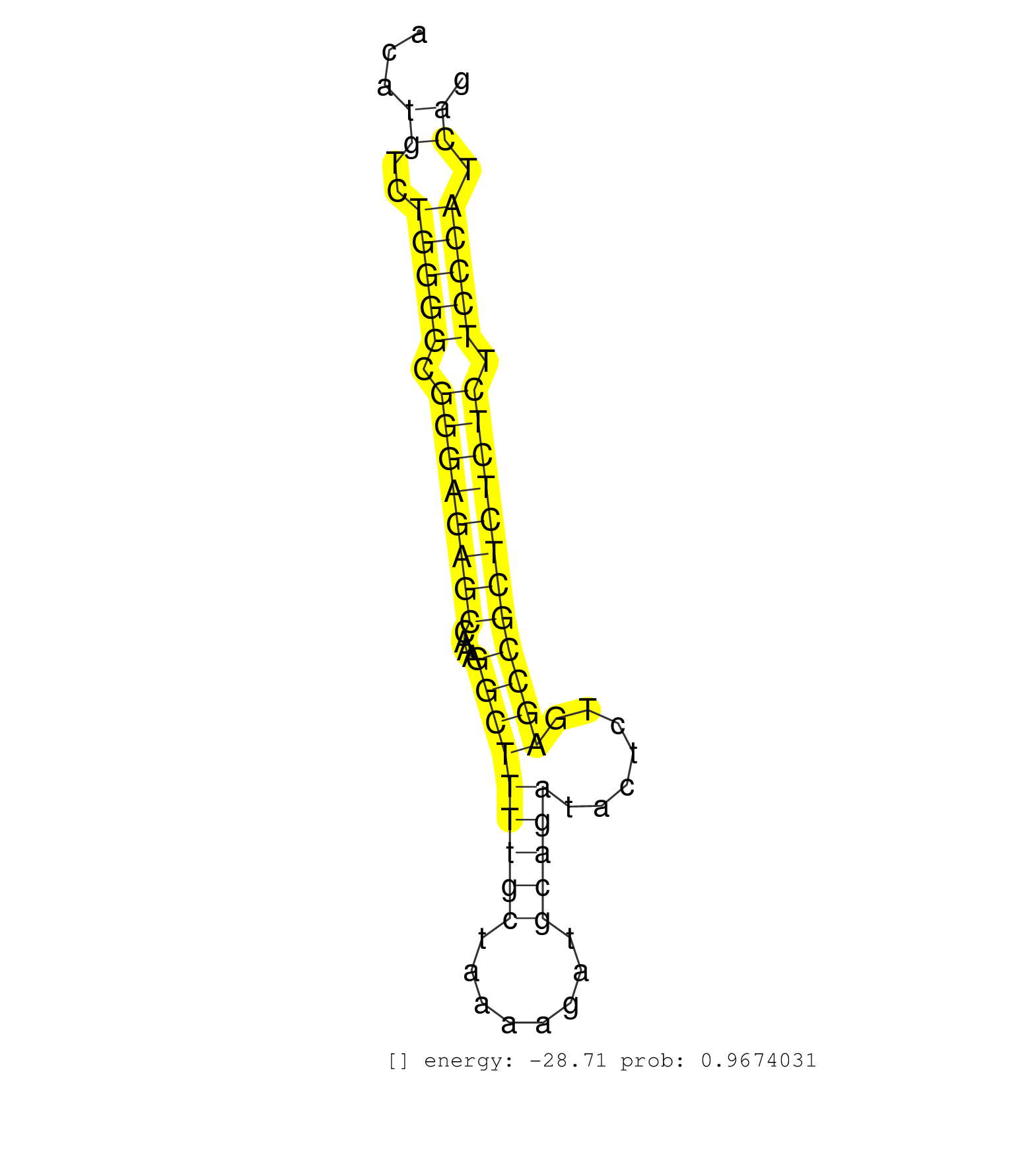

| Gene: Epas1 | ID: uc008duj.1_intron_7_0_chr17_87223168_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| GGAGCGAGCGAACAGCCCACAGGTTGAGGCTAACCTCACCGTCTCTGCTTCTGTGGCTCACAGAATTGGATTGTTTGTGTTTCTGCCTAACACTTCTCTGAGAGGTTTATCTACCAGGCTTCTGACATGTCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTCCCATCAGTGAGATCGAGAAGAACGACGTGGTGTTCTCCATGGACCAGACCGAATCCC ...............................................................................................................................((..(((((.((((((((....(((((((((........))))).......)))))))))))).))))).))................................................... ............................................................................................................................125........................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................CGACGTGGTGTTCTCCATGGACCAG.......... | 25 | 1 | 10.00 | 10.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTGCTAAAAGATGCAGATACTCTGA....................................................................... | 25 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCGCTCTCTCTTCCCATC.................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTT............................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................AAAGATGCAGATACTCTGAGCC.................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................CCATGGACCAGACCGAAT... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCCCATCAGTGAGATCGAGAAGAAC.................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TCAGTGAGATCGAGAAGAACGACGTG............................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGTGAGATCGAGAAGAACGACGTG............................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGG................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TGAGATCGAGAAGAACGACGTGGTG......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGC.................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................TCCCATCAGTGAGATCGAGAAGAACGAC............................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................AGATACTCTGAGCCGCcggg.............................................................. | 20 | cggg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCCCATCAGTGAGATCGAGAAGAACG................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................TAACCTCACCGTCTCTGCTTCTGTGG.................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCGCTCTCTCTTCCCAT..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GAGATCGAGAAGAACGACGTG............................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| GGAGCGAGCGAACAGCCCACAGGTTGAGGCTAACCTCACCGTCTCTGCTTCTGTGGCTCACAGAATTGGATTGTTTGTGTTTCTGCCTAACACTTCTCTGAGAGGTTTATCTACCAGGCTTCTGACATGTCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTCCCATCAGTGAGATCGAGAAGAACGACGTGGTGTTCTCCATGGACCAGACCGAATCCC ...............................................................................................................................((..(((((.((((((((....(((((((((........))))).......)))))))))))).))))).))................................................... ............................................................................................................................125........................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|