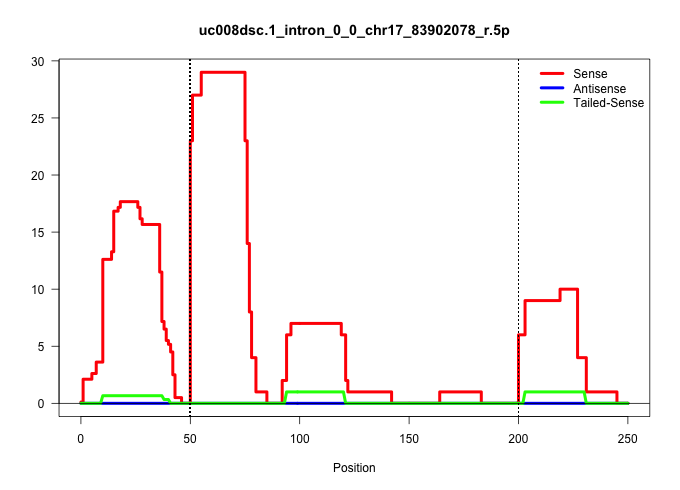
| Gene: Cox7a2l | ID: uc008dsc.1_intron_0_0_chr17_83902078_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(27) TESTES |
| ATGATTATTCTGGGAAGAACAAAGTTCCAGAGCTGCAGAAGTTTTTCCAGGTGAGTGAAGATGGGAACACAGAACTGTGAAATGCACACACATGTGTGTTGGCTGGGAGCTTCTAAAACTGACTTGACTTTAGTGTTGTTTTCTCCCCAGCCTCCTTTTCTTAAGGAGAACTCTTTACTTCAGTTTGGTCCGTTGGCTAATAATGGAACCTTTGTACAGGTAGATGCTGGTAACAGGGGCGTGGCTAATG ..................................................((.(((((((.(((...((((....))))....((((((....))))))...)))....))))....))).))............................................................................................................................... ..................................................51........................................................................125........................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTGAAGATGGGAACACAGAACT.............................................................................................................................................................................. | 26 | 1 | 8.00 | 8.00 | 2.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TAATGGAACCTTTGTACAGGTAGATGC....................... | 27 | 1 | 6.00 | 6.00 | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGAAGATGGGAACACAGAAC............................................................................................................................................................................... | 25 | 1 | 6.00 | 6.00 | 2.00 | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGAAGATGGGAACACAGAACTG............................................................................................................................................................................. | 27 | 1 | 5.00 | 5.00 | 2.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGGAAGAACAAAGTTCCAGAGCTGC...................................................................................................................................................................................................................... | 26 | 3 | 5.00 | 5.00 | 1.00 | 1.33 | - | - | - | - | - | 2.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| ..........TGGGAAGAACAAAGTTCCAGAGCTGCA..................................................................................................................................................................................................................... | 27 | 3 | 4.33 | 4.33 | - | - | 0.33 | 0.33 | 1.33 | 0.67 | - | - | - | 0.33 | - | - | - | - | 0.67 | - | - | - | - | 0.33 | - | - | - | - | - | 0.33 | - |
| ..................................................GTGAGTGAAGATGGGAACACAGAACTGT............................................................................................................................................................................ | 28 | 1 | 4.00 | 4.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGAACCTTTGTACAGGTAGATGCTGGT................... | 28 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTGGCTGGGAGCTTCTAAAACTG................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGAAGATGGGAACACAGAACTGTGA.......................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGAACAAAGTTCCAGAGCTGCAGAAGT................................................................................................................................................................................................................ | 27 | 3 | 1.67 | 1.67 | - | - | 0.67 | 0.33 | - | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGAACAAAGTTCCAGAGCTGCAGAAGTT............................................................................................................................................................................................................... | 28 | 3 | 1.67 | 1.67 | - | - | 0.33 | 0.67 | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGGAAGAACAAAGTTCCAGAGCTGCAGA................................................................................................................................................................................................................... | 29 | 3 | 1.33 | 1.33 | 0.33 | - | - | 0.33 | - | - | - | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GTAGATGCTGGTAACAGGGGCGTGGC..... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGTGTGTTGGCTGGGAGCTTCTAAAACTG................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGAAGATGGGAACACAGAACT.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................AGGTAGATGCTGGTAcaga.............. | 19 | caga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGAACCTTTGTACAGGTAGATGCTGGg................... | 28 | g | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTGGCTGGGAGCTTCTAAAAC................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................ACTGACTTGACTTTAGTGTTGTTTT............................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTGGCTGGGAGCTTCTAAAAatg................................................................................................................................. | 27 | atg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGAGAACTCTTTACTTCAG................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGAAGATGGGAACACAGAACTGTGAAATGC..................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TTCTGGGAAGAACAAAGTTCCAGAGCTGC...................................................................................................................................................................................................................... | 29 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ............................................................................................TGTGTGTTGGCTGGGAGCTTCTAAA..................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGATTATTCTGGGAAGAACAAAGTTC............................................................................................................................................................................................................................... | 26 | 2 | 1.00 | 1.00 | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGAAGATGGGAACACAGAACTG............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGTTGGCTGGGAGCTTCTAAAACTGA................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGAAGATGGGAACACAGAACTGTGA.......................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GAAGAACAAAGTTCCAGAGCTGCAGA................................................................................................................................................................................................................... | 26 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGGAAGAACAAAGTTCCAGAGCTGCAG.................................................................................................................................................................................................................... | 28 | 3 | 0.67 | 0.67 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| .TGATTATTCTGGGAAGAACAAAGTTCC.............................................................................................................................................................................................................................. | 27 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................ACAAAGTTCCAGAGCTGCAGAAGTTTTT............................................................................................................................................................................................................ | 28 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TATTCTGGGAAGAACAAAGTTCCAGAGCTGC...................................................................................................................................................................................................................... | 31 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGATTATTCTGGGAAGAACAAAGTT................................................................................................................................................................................................................................ | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGGAAGAACAAAGTTCCAGA........................................................................................................................................................................................................................... | 21 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GAAGAACAAAGTTCCAGAGCTGCAGAA.................................................................................................................................................................................................................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGGAAGAACAAAGTTCCAGAGCTGCAGAA.................................................................................................................................................................................................................. | 30 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ..........TGGGAAGAACAAAGTTCCAGAGCTGCAGAAt................................................................................................................................................................................................................. | 31 | t | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| ..............AAGAACAAAGTTCCAGAGCTGCAGAAG................................................................................................................................................................................................................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ..............AAGAACAAAGTTCCAGAGCTGCAGAAGT................................................................................................................................................................................................................ | 28 | 3 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGAACAAAGTTCCAGAGCTGCAGAAG................................................................................................................................................................................................................. | 26 | 3 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................AACAAAGTTCCAGAGCTGCAGAAGTT............................................................................................................................................................................................................... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGGAAGAACAAAGTTCCAGAGCTGCta.................................................................................................................................................................................................................... | 28 | ta | 0.33 | 5.00 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............AAGAACAAAGTTCCAGAGCTGCAG.................................................................................................................................................................................................................... | 24 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - |
| .........CTGGGAAGAACAAAGTTC............................................................................................................................................................................................................................... | 18 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - |
| ATGATTATTCTGGGAAGAACAAAGTTCCAGAGCTGCAGAAGTTTTTCCAGGTGAGTGAAGATGGGAACACAGAACTGTGAAATGCACACACATGTGTGTTGGCTGGGAGCTTCTAAAACTGACTTGACTTTAGTGTTGTTTTCTCCCCAGCCTCCTTTTCTTAAGGAGAACTCTTTACTTCAGTTTGGTCCGTTGGCTAATAATGGAACCTTTGTACAGGTAGATGCTGGTAACAGGGGCGTGGCTAATG ..................................................((.(((((((.(((...((((....))))....((((((....))))))...)))....))))....))).))............................................................................................................................... ..................................................51........................................................................125........................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................TTTCTCCCCAGCCTCttt................................................................................................ | 18 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................TTCTAAAACTGACTTGtgc............................................................................................................................ | 19 | tgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |