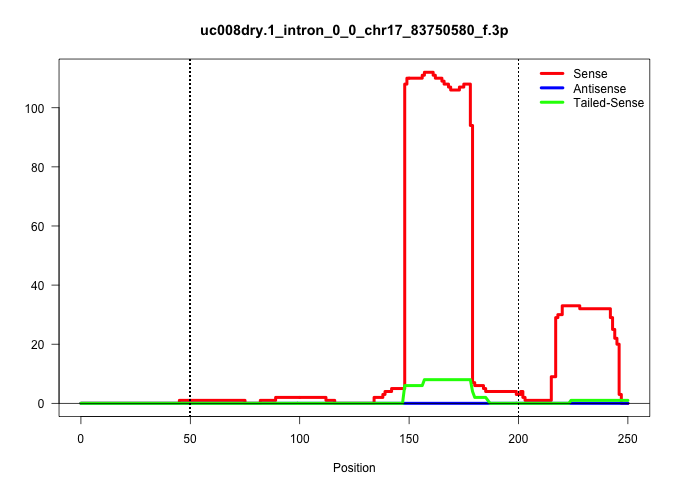
| Gene: Eml4 | ID: uc008dry.1_intron_0_0_chr17_83750580_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| ATCATAAATTATATTTGTGACCCCATCAATGGGAAAACAAGAAATTACAGTGTTCCCTTTTCAAAAGATTTATATAAAGTATTAAAATTCTAAAGGTACTAAACTGTATAGCCATGTTCACTTTAATACAATGCTGGGTTGCCAGATTGAGGATATCTAGAATACAGTGTATTTCTGCAAAGGTGACATGTTTCTTCAAGTTCAGGAAACAAGCCTGTGATGGATTGTAATAGAAGCAAAATGAACAGAG ......................................................................................................................................................((((....((((....((((...(((....)))..))))..))))....))))............................................... ...............................................................................................................................................144...........................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTCTGCA....................................................................... | 31 | 1 | 85.00 | 85.00 | 39.00 | 21.00 | 12.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGATGGATTGTAATAGAAGCAAAATGAAC.... | 29 | 1 | 17.00 | 17.00 | - | - | - | - | 7.00 | 3.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - |
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTCTGC........................................................................ | 30 | 1 | 15.00 | 15.00 | 6.00 | 3.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| .......................................................................................................................................................................................................................TGTGATGGATTGTAATAGAAGCAAAAT........ | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTC........................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................AGGATATCTAGAATACAGTGTATTTCTGCA....................................................................... | 30 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGCAAAGGTGACATGTTTCTTCAAGTT................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGGATTGTAATAGAAGCAAAATGAACA... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGATGGATTGTAATAGAAGCAAAATG....... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGTGATGGATTGTAATAGAAGCAAAATG....... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTCTGCAt...................................................................... | 32 | t | 2.00 | 85.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGATGGATTGTAATAGAAGCAAAATGA...... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTCTGCt....................................................................... | 31 | t | 1.00 | 15.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TCAGGAAACAAGCCTGTGATGGATTGT...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGCCAGATTGAGGATATCTAGAATACAGT.................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TTGTAATAGAAGCAAAATGAACAGAGtata | 30 | tata | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGTTGCCAGATTGAGGATATCTAGAA........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CTAAAGGTACTAAACTGTATAGCCATG...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TAAAATTCTAAAGGTACTAAACTGTATAGC.......................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGCAAAGGTGACATGTTTCTTCAAGTTC............................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTCTtca....................................................................... | 31 | tca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GATGGATTGTAATAGAAGCAAAATGAACA... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TAGAATACAGTGTATTTCTGCAAAGGTGtt............................................................... | 30 | tt | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TAGAATACAGTGTATTTCTGCAAAGGTGt................................................................ | 29 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTGCCAGATTGAGGATATCTAGAATAC..................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGGATTGTAATAGAAGCAAAATGAA..... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTCTGCAA...................................................................... | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TACAGTGTTCCCTTTTCAAAAGATTTATAT............................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TCTGCAAAGGTGACATGTTTCTTCAA................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGTGATGGATTGTAATAGAAGCAAAATGAA..... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTCcgca....................................................................... | 31 | cgca | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGTTGCCAGATTGAGGATATCTAGA......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGTGATGGATTGTAATAGAAGCAAAATGA...... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CAGATTGAGGATATCTAGAATACAGTG................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGGATATCTAGAATACAGTGTATTTCTGta....................................................................... | 31 | ta | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGTGATGGATTGTAATAGAAGCAAAATGAAC.... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TAGAATACAGTGTATTTCTGCAAAGGTG................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGGATATCTAGAATACA.................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CTAGAATACAGTGTATTTCTGCAAAGGT.................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ATCATAAATTATATTTGTGACCCCATCAATGGGAAAACAAGAAATTACAGTGTTCCCTTTTCAAAAGATTTATATAAAGTATTAAAATTCTAAAGGTACTAAACTGTATAGCCATGTTCACTTTAATACAATGCTGGGTTGCCAGATTGAGGATATCTAGAATACAGTGTATTTCTGCAAAGGTGACATGTTTCTTCAAGTTCAGGAAACAAGCCTGTGATGGATTGTAATAGAAGCAAAATGAACAGAG ......................................................................................................................................................((((....((((....((((...(((....)))..))))..))))....))))............................................... ...............................................................................................................................................144...........................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|