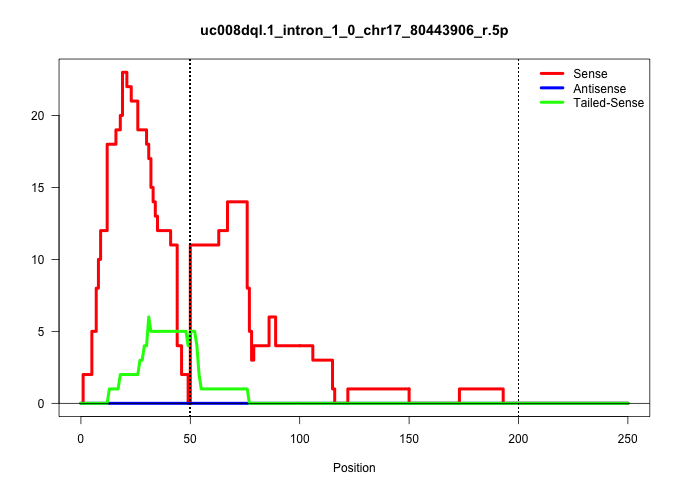
| Gene: Hnrpll | ID: uc008dql.1_intron_1_0_chr17_80443906_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(22) TESTES |
| GAATGACAATGACAGTTGGGACTACACTAAACCTTATTTGGGAAGACGAGGTAGGTATTTTACGAATTTTAGAATGTACTGAGATCTGCTGAGAAACTTTACAAGCAGTCGCATTCTTTTTTTTCTGAGATGAGCTAGTTAATTAAAATGAAAATCTTGTTGAATGCTTTTGTTACAAAAGTCCGTGTAATGCTGGAAATTAACCTTGGACATTAGTGCCCCCTACTGGTACCTGCTGAGCTTATTTATA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............CAGTTGGGACTACACTAAACCTTATTTGGGAA.............................................................................................................................................................................................................. | 32 | 1 | 6.00 | 6.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATTTTACGAATTTTAGAATG.............................................................................................................................................................................. | 26 | 1 | 6.00 | 6.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAGGTATTTTACGAATTTTAGAATGT............................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TTTAGAATGTACTGAGATCTGC................................................................................................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATTTTACGAATTTTAGAATGTA............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GACTACACTAAACCTTATTTGGGAAGACGA......................................................................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......AATGACAGTTGGGACTACA................................................................................................................................................................................................................................ | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGCTGAGAAACTTTACAAGCAGTCGCATT....................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....ACAATGACAGTTGGGACTACACTAAAC.......................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................TTCTGAGATGAGCTAGTTAATTAAAATG.................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................TGAGAAACTTTACAAGCAGTCGCATTC...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .AATGACAATGACAGTTGGGACT................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............AGTTGGGACTACACTAAAt.......................................................................................................................................................................................................................... | 19 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................CCTTATTTGGGAAGACGAGGTtg.................................................................................................................................................................................................... | 23 | tg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................TGGGACTACACTAAACCTTATTTGGGAAGA............................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....ACAATGACAGTTGGGACTACACTAA............................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .AATGACAATGACAGTTGGGA..................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GAATTTTAGAATGTACTGAGATCTGC................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TAAACCTTATTTGGGAAGACGAGGTt..................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........ATGACAGTTGGGACTACACTAAA........................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGACAGTTGGGACTACACTAAACCTTATTTGG................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................AACCTTATTTGGGAAGACGAGGTtg.................................................................................................................................................................................................... | 25 | tg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ATGACAGTTGGGACTACACTAAACCTT....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GGACTACACTAAACCTTATTTGGGAA.............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TACAAAAGTCCGTGTAATGC......................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................CCTTATTTGGGAAGACGAGGTtgt................................................................................................................................................................................................... | 24 | tgt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TGAGATCTGCTGAGAAACTTTACAAGC................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GACTACACTAAACCTTATTTGGGAAGA............................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......AATGACAGTTGGGACTACACTAAACC......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........TGACAGTTGGGACTACACTAAACCT........................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................GGACTACACTAAACCTTATTTGGGAAGACGt......................................................................................................................................................................................................... | 31 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATTTTACGAATTTTAGAATGa............................................................................................................................................................................. | 27 | a | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GAATGACAATGACAGTTGGGACTACACTAAACCTTATTTGGGAAGACGAGGTAGGTATTTTACGAATTTTAGAATGTACTGAGATCTGCTGAGAAACTTTACAAGCAGTCGCATTCTTTTTTTTCTGAGATGAGCTAGTTAATTAAAATGAAAATCTTGTTGAATGCTTTTGTTACAAAAGTCCGTGTAATGCTGGAAATTAACCTTGGACATTAGTGCCCCCTACTGGTACCTGCTGAGCTTATTTATA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|