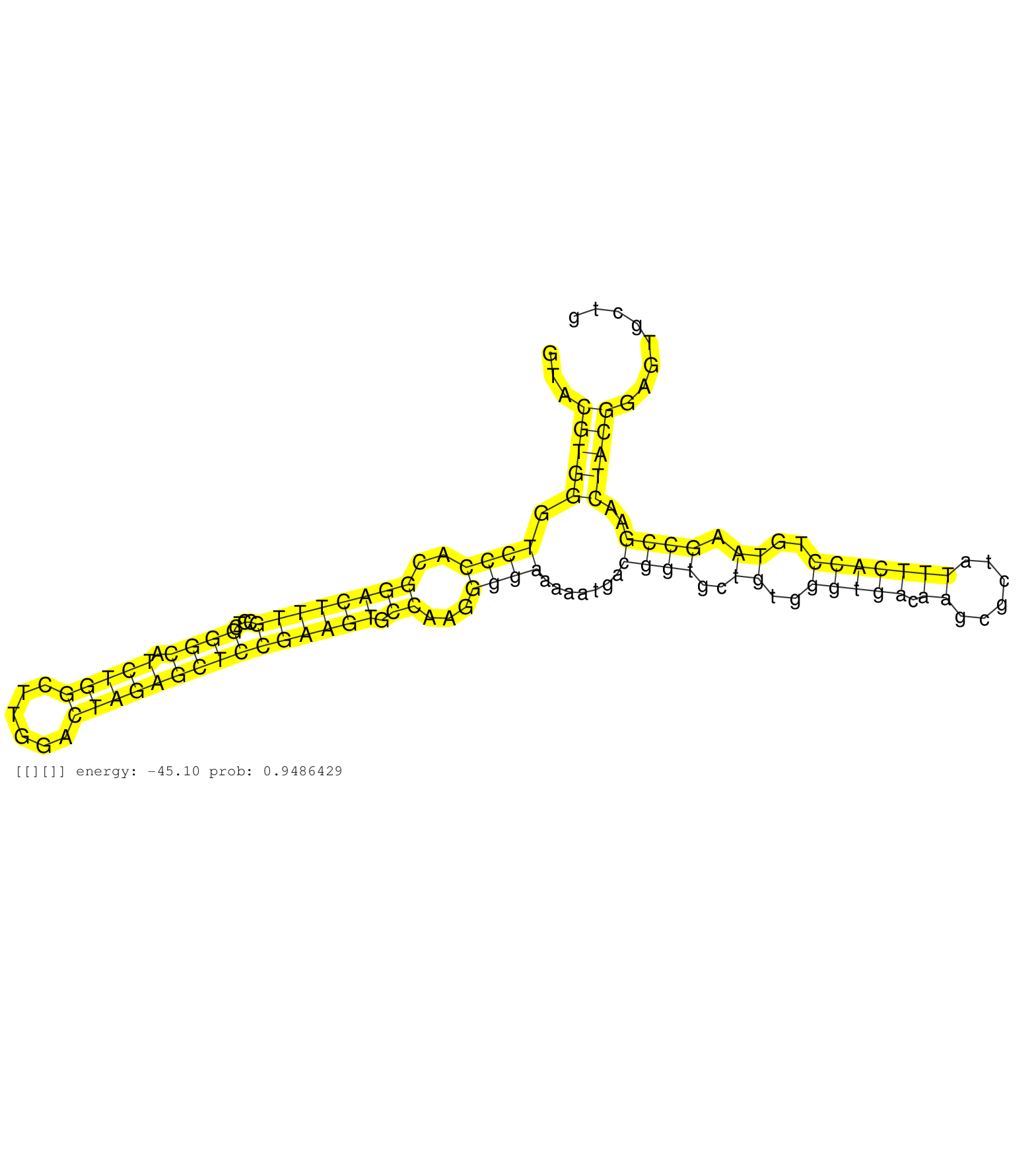

| Gene: Clip4 | ID: uc008dmy.1_intron_15_0_chr17_72205694_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(9) TESTES |
| GAGGGGTCTCAGGTCCTGCTCACAAGCTCTAACGAAATGGCCACGGTTAGGTACGTGGGTCCCACGGACTTTGCCTCGGGCATCTGGCTTGGACTAGAGCTCCGAAGTGCCAAGGGGAAAAATGACGGTGCTGTGGGTGACAAGCGCTATTTCACCTGTAAGCCGAACTACGGAGTGCTGGTTAGACCAAGCCGAGTGACCTACCGCGGGATTAGTGGGTCAAAGCTCATTGATGAGAATTGTTGAGTTA .....................................................(((((.((((..((((((((....((((.(((((......))))))))))))))).))...)))).......((((..((..(((((.((......)))))))..)).))))..))))).............................................................................. ..................................................51...............................................................................................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTACGTGGGTCCCACGGACTTTGCCTCGGGCATCTGGCTTGGACTAGAGCTC.................................................................................................................................................... | 52 | 1 | 21.00 | 21.00 | 21.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................AAAAATGACGGTGCTGTGGGTGtta............................................................................................................ | 25 | tta | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTTCACCTGTAAGCCGAACTACGGAGT.......................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................GGGCATCTGGCTTGGACTA.......................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................AAATGACGGTGCTGTGGG................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................GGAAAAATGACGGTGCTGTGG.................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................TGACAAGCGCTATTTCACCTGTAAGCC...................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................AAAATGACGGTGCTGTGGGTGACAA........................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................AAGCCGAGTGACCTACCGCGGGATTAGTGG................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| GAGGGGTCTCAGGTCCTGCTCACAAGCTCTAACGAAATGGCCACGGTTAGGTACGTGGGTCCCACGGACTTTGCCTCGGGCATCTGGCTTGGACTAGAGCTCCGAAGTGCCAAGGGGAAAAATGACGGTGCTGTGGGTGACAAGCGCTATTTCACCTGTAAGCCGAACTACGGAGTGCTGGTTAGACCAAGCCGAGTGACCTACCGCGGGATTAGTGGGTCAAAGCTCATTGATGAGAATTGTTGAGTTA .....................................................(((((.((((..((((((((....((((.(((((......))))))))))))))).))...)))).......((((..((..(((((.((......)))))))..)).))))..))))).............................................................................. ..................................................51...............................................................................................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................CCAAGGGGAAAAATtggt............................................................................................................................... | 18 | tggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| ...................................ATGGCCACGGTTAGGTAc..................................................................................................................................................................................................... | 18 | c | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - |