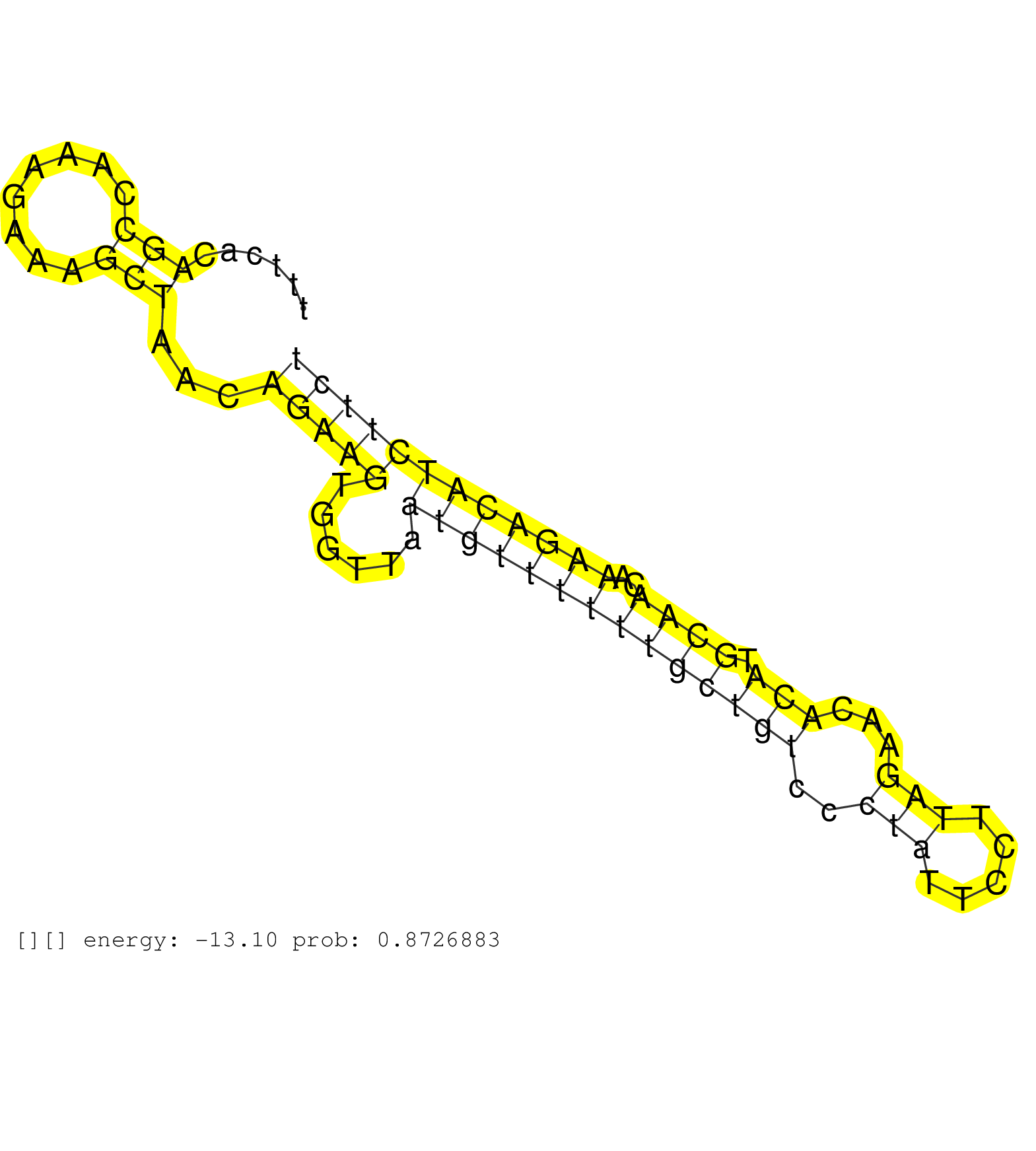

| Gene: AK082862 | ID: uc008dln.1_intron_3_0_chr17_71154489_r.3p | SPECIES: mm9 |
 |
 |
|
(6) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| ATGCTGTGCTATGGAGGGCTCTAAGCAACACAGTTGAGCGACCATGGGCTTCTGAACCCTTTCCTCTCCTGACTTGTTTTCCCCCAGGTCTTTTTTCACAGCCAAAGAAAGCTAACAGAAGTGGTTAATGTTTTTTGCTGTCCCTATTCCTTAGAACACATGCAACAAAGACATCTTCTAAAATAACCCACTCTGCTCAGTTCTCCAGGCATGCCCTTGAACGTGGCTTATATCACCTGTCCATCTGATA ...................................................................................................(((........)))...(((((......((((((((((((((..(((.....)))...))).))))..))))))))))))....................................................................... .............................................................................................94...................................................................................179..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TTCCTTAGAACACATGCAACAAAGACATC........................................................................... | 29 | 1 | 20.00 | 20.00 | 9.00 | 4.00 | - | 1.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TAGAACACATGCAACAAAGACATCTTC........................................................................ | 27 | 1 | 11.00 | 11.00 | 3.00 | - | 3.00 | 1.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TATTCCTTAGAACACATGCAACAAAGACA............................................................................. | 29 | 1 | 10.00 | 10.00 | 3.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................TATTCCTTAGAACACATGCAACAAAGAC.............................................................................. | 28 | 1 | 10.00 | 10.00 | 2.00 | 3.00 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TATTCCTTAGAACACATGCAACAAAGACATC........................................................................... | 31 | 1 | 7.00 | 7.00 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................TTCCTTAGAACACATGCAACAAAGACA............................................................................. | 27 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TAGAACACATGCAACAAAGACATCTTCT....................................................................... | 28 | 1 | 5.00 | 5.00 | 1.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TAGAACACATGCAACAAAGACATCTTCTA...................................................................... | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 |
| ........................................................................................................................................................................................................TTCTCCAGGCATGCCCTTGAACGTGGCTT..................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTCCTTAGAACACATGCAACAAAGACAT............................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - |
| ......................................................................................................................................................TTAGAACACATGCAACAAAGACATCT.......................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TTAGAACACATGCAACAAAGACATC........................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TGCTCAGTTCTCCAGGCATGCCCTTGA.............................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TAACCCACTCTGCTCAGTTCTCCAGGCATa..................................... | 30 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TAGAACACATGCAACAAAGACATC........................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TAAGCAACACAGTTGAGCGACCATGGG.......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGGAGGGCTCTAAGCAACACAGTTGA..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TGAGCGACCATGGGCTTCTGAACCCTTTC........................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................TGAGCGACCATGGGCTTCTGAACCC............................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTCCTTAGAACACATGCAACAAAGACATCT.......................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CAGCCAAAGAAAGCTAACAGAAGTGGTT............................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................TGAGCGACCATGGGCgggc..................................................................................................................................................................................................... | 19 | gggc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGAACGTGGCTTATATCAC.............. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TAAGCAACACAGTTGAGCGACCA.............................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................TCCTTAGAACACATGCAACAAAGACATCT.......................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCCTTAGAACACATGCAACAAAGACA............................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TATTCCTTAGAACACATGCAACAAAGACAT............................................................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TTAGAACACATGCAACAAAGACATCTT......................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TCCAGGCATGCCCTTGAACGTGGCTT..................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .TGCTGTGCTATGGAGGGCTCTAAGCAAC............................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCCTTAGAACACATGCAACAAAGACATC........................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TATTCCTTAGAACACATGCAACAAAGACATCT.......................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ATGCTGTGCTATGGAGGGCTCTAAGCAACACAGTTGAGCGACCATGGGCTTCTGAACCCTTTCCTCTCCTGACTTGTTTTCCCCCAGGTCTTTTTTCACAGCCAAAGAAAGCTAACAGAAGTGGTTAATGTTTTTTGCTGTCCCTATTCCTTAGAACACATGCAACAAAGACATCTTCTAAAATAACCCACTCTGCTCAGTTCTCCAGGCATGCCCTTGAACGTGGCTTATATCACCTGTCCATCTGATA ...................................................................................................(((........)))...(((((......((((((((((((((..(((.....)))...))).))))..))))))))))))....................................................................... .............................................................................................94...................................................................................179..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................GAGCGACCATGGcgg........................................................................................................................................................................................................... | 15 | cgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |