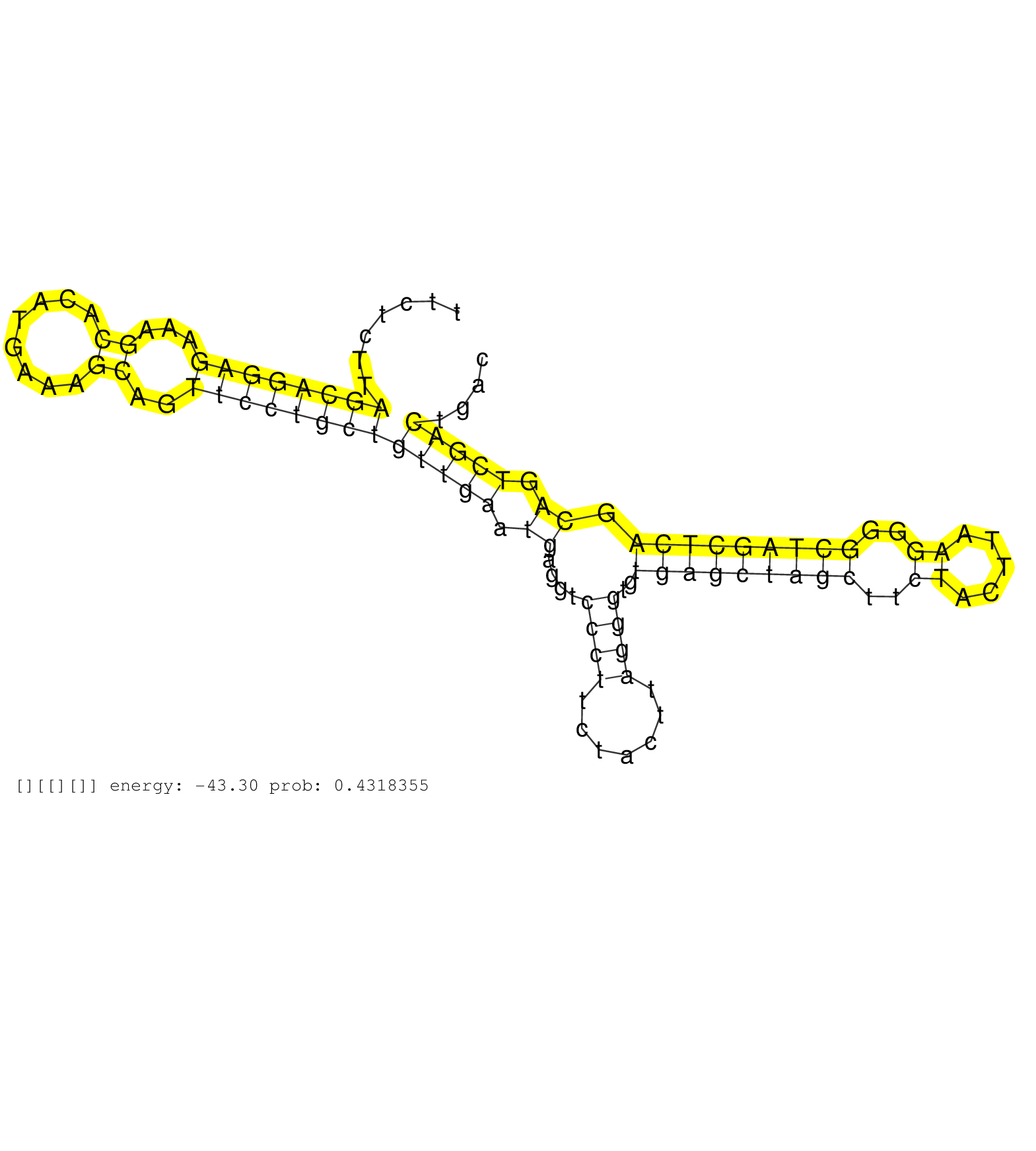
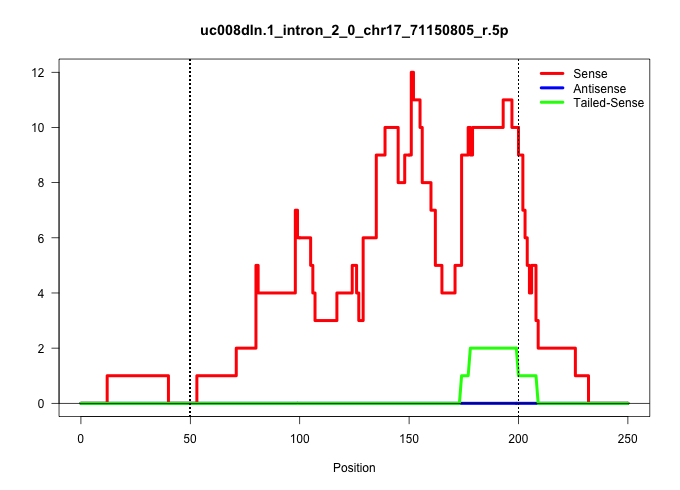
| Gene: AK082862 | ID: uc008dln.1_intron_2_0_chr17_71150805_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| TGGAGGAAAGCATAGTCTCCGAAGCCTTGTTTTGAGCACCTCTTGGGGAGGTGTGTGGATCTTTCCTCTCATTTCCTTTCTACATGCTGGTATTTCTCTTAGCAGGAGAAAGCACATGAAAGCAGTTCCTGCTGTTGAATGAGGTCCCTTCTACTTAGGGTGTGAGCTAGCTTCTACTTAAGGGGCTAGCTCAGCAGTCGACTGACTCACAACTGCCTATGGTTCCAGCTCCAGAATGATCAAGCTCTTC ....................................................................................................((((((((...((........))..))))))))(((((.((....((((.......))))..(((((((((..((.....))..))))))))).)).)))))................................................ .............................................................................................94..............................................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................TACTTAAGGGGCTAGCTCAGCAGTCGAC................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTAGCAGGAGAAAGCACATGAAAGCAGT............................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................TACTTAGGGTGTGAGCTAGCTTCTACT........................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGCTGTTGAATGAGGTCCCTTCTACTT.............................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................TGAATGAGGTCCCTTCTACTTAGGGTG........................................................................................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................AAGGGGCTAGCTCAGCAGTCGACTGACTC.......................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................TGAATGAGGTCCCTTCTACTTAGGG.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TACTTAGGGTGTGAGCTAGCTTCTACTT....................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TAGTCTCCGAAGCCTTGTTTTGAGCACC.................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTAGCAGGAGAAAGCACATGAAAGCAGTT........................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TACTTAAGGGGCTAGCTCAGCAGTCGACTGA............................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................GAAAGCAGTTCCTGCTGTTGAATGAGGT......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTCTACTTAAGGGGCTAGCTCAGCAG..................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TACTTAAGGGGCTAGCTCAGCAGTCGACT............................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCCTGCTGTTGAATGAGGTCCCTTCT.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TTTCCTTTCTACATGCTGGTATTTCTCT....................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TACATGCTGGTATTTCTCTTAGCAGG................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTCCAGCTCCAGAATGATCAAGCTCT.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TAAGGGGCTAGCTCAGCAGTCGACTGACTCt......................................... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TAAGGGGCTAGCTCAGCAGTCGACTGACTCA......................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................GCAGTCGACTGACTCACAACTGCCTATGGTTCC........................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................CTTGTTTTGAGCACCTagga............................................................................................................................................................................................................. | 20 | agga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................TTAAGGGGCTAGCTCAGCAGTCGACTG.............................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GTTCCTGCTGTTGAATGAGGT......................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................TGTGGATCTTTCCTCTCATTTCCTTTCT......................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGCTGTTGAATGAGGTCCCTTCTACT............................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAGGTCCCTTCTACTTAGGGTGTGA..................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TACTTAAGGGGCTAGCTCAGCAGTCG.................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TCCGAAGCCTTGTTTTGAGCACCTCc............................................................................................................................................................................................................... | 26 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................TACATGCTGGTATTTCTCTTAGCAG................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCACAACTGCCTATGGTTCCAGCTCC.................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TTCTACTTAGGGTGTGAGCTAGCTTC............................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TACTTAAGGGGCTAGCTCAGCAGTtg.................................................. | 26 | tg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................TACATGCTGGTATTTCTCTTAGCAGGA............................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| TGGAGGAAAGCATAGTCTCCGAAGCCTTGTTTTGAGCACCTCTTGGGGAGGTGTGTGGATCTTTCCTCTCATTTCCTTTCTACATGCTGGTATTTCTCTTAGCAGGAGAAAGCACATGAAAGCAGTTCCTGCTGTTGAATGAGGTCCCTTCTACTTAGGGTGTGAGCTAGCTTCTACTTAAGGGGCTAGCTCAGCAGTCGACTGACTCACAACTGCCTATGGTTCCAGCTCCAGAATGATCAAGCTCTTC ....................................................................................................((((((((...((........))..))))))))(((((.((....((((.......))))..(((((((((..((.....))..))))))))).)).)))))................................................ .............................................................................................94..............................................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................CGAAGCCTTGTTTTGAGct...................................................................................................................................................................................................................... | 19 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |