
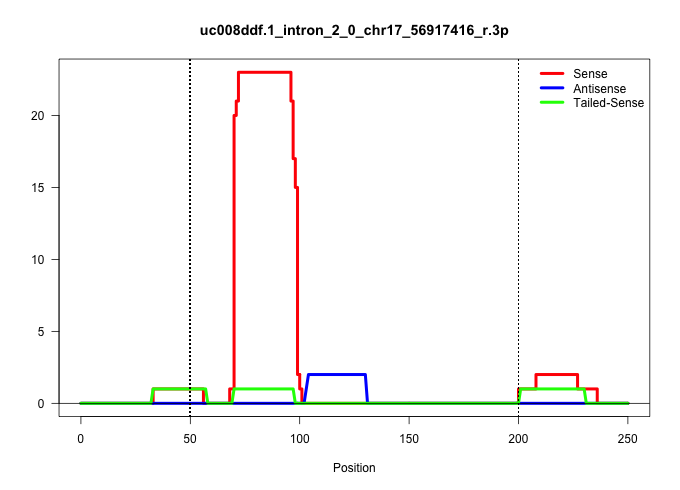
| Gene: Rfx2 | ID: uc008ddf.1_intron_2_0_chr17_56917416_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(14) TESTES |
| TGCGTTGTACCCACCTGTATCCCCCTATGTCCCTGTGTATGTCTGGGAATGGGTGGCCATCCCCTGCCTCTGTGGACCTGATGACCTCAAGCAGGCAGCCATTGCAGTGCTTGCAGGCTCAGAGCATCCCAGAGGAGCCAGGCCAGTACCCCAGGCTCAGTTCCAGTGATGGTGCCCATTCTCTGTCCCTGTGTCCCCAGCTCCATGGTGATCCGTGACCTGACCCTGCGCAGTGCTGCCAGCTTCGGTT ................................................................................................................(((((..(((((((((.((..(((((..((((.........))))..)))))..))..)))))......))))..))))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TGTGGACCTGATGACCTCAAGCAGGCAGC....................................................................................................................................................... | 29 | 1 | 13.00 | 13.00 | 4.00 | 5.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGTGGACCTGATGACCTCAAGCAGGCA......................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................TGTGGACCTGATGACCTCAAGCAGGCAG........................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| .................................TGTGTATGTCTGGGAATGGGTGGCa................................................................................................................................................................................................ | 25 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGGACCTGATGACCTCAAGCAGGCAGCCA..................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGTGGACCTGATGACCTCAAGCAGGCAt........................................................................................................................................................ | 28 | t | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................TGTGTATGTCTGGGAATGGGTGG.................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................CTCCATGGTGATCCGTGACCTGACCCT....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................TGTGGACCTGATGACCTCAAGCAGGC.......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GTGGACCTGATGACCTCAAGCAGGCAG........................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGGACCTGATGACCTCAAGCAGGCAGCC...................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TGATCCGTGACCTGACCCTGCGCAGTGC.............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................TCTGTGGACCTGATGACCTCAAGCAGGC.......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TCCATGGTGATCCGTGACCTGACCCTGCGt................... | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| TGCGTTGTACCCACCTGTATCCCCCTATGTCCCTGTGTATGTCTGGGAATGGGTGGCCATCCCCTGCCTCTGTGGACCTGATGACCTCAAGCAGGCAGCCATTGCAGTGCTTGCAGGCTCAGAGCATCCCAGAGGAGCCAGGCCAGTACCCCAGGCTCAGTTCCAGTGATGGTGCCCATTCTCTGTCCCTGTGTCCCCAGCTCCATGGTGATCCGTGACCTGACCCTGCGCAGTGCTGCCAGCTTCGGTT ................................................................................................................(((((..(((((((((.((..(((((..((((.........))))..)))))..))..)))))......))))..))))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................GCAGTGCTTGCAGGCTCAGAGCATCCCA....................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................CAGTGCTTGCAGGCTCAGAGCATCCCA....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................................................CTGTCCCTGTGTCCCCAGCTCCA............................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTCTGTCCCTGTGTCCCCAGCTCCA............................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |