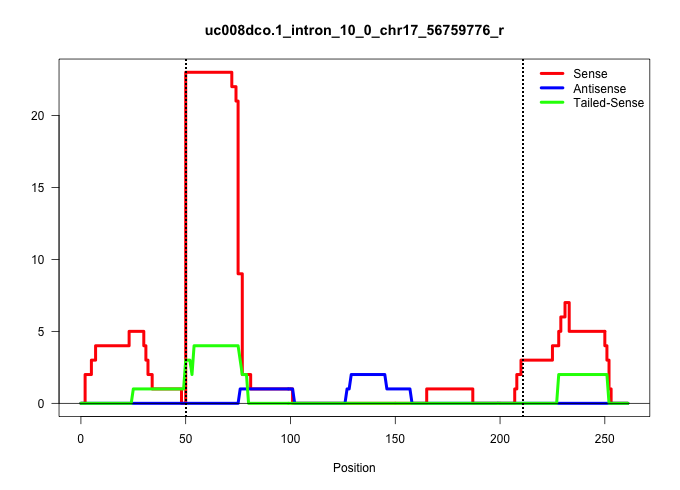
| Gene: Lonp1 | ID: uc008dco.1_intron_10_0_chr17_56759776_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| GAGATTGTGAAGACCATTCGGGATATCATCGCCTTGAACCCTCTGTACAGGTCAGTGTCTGCAGGGCTGAGGAGGACAGGGACGGTCAGTGACAGCTGCTTGACCCCAGTGCACCACACTGCACACTGCCAGAGCTATCTATCACTGAAGCCAGAGTATGTGTAAGGGGCATAGGACAGCACTCTAAGGCACTGGCACTGAACCCCTACAGAGAGTCCGTGCTGCAGATGATGCAGGCAGGCCAGCGTGTGGTGGACAACC ..................................................(((..(((((.((....))....)))))..)))..(((((((.((..((((.......(((((......))))).......))))...)).))))))).((((((((.(((((.....))))).......)))))..)))....................................................................... ..................................................51..........................................................................................................................................191.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTCAGTGTCTGCAGGGCTGAGGAGG.......................................................................................................................................................................................... | 25 | 1 | 41.00 | 41.00 | 17.00 | 1.00 | 3.00 | 4.00 | 2.00 | 1.00 | 4.00 | 2.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ..................................................GTCAGTGTCTGCAGGGCTGAGGAGGAC........................................................................................................................................................................................ | 27 | 1 | 7.00 | 7.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................CATCGCCTTGAACCCTCTGTACAGag................................................................................................................................................................................................................. | 26 | ag | 6.00 | 0.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............CCATTCGGGATATCATCGCCTT.................................................................................................................................................................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................TGATGCAGGCAGGCCAGCGTGTGt......... | 24 | t | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGTCTGCAGGGCTGAGGAGGACAGa..................................................................................................................................................................................... | 26 | a | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................TGATGCAGGCAGGCCAGCGTGTGG......... | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGAAGACCATTCGGGATATCATCGCCT................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TATCATCGCCTTGAACCCTCTGTAC..................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CAGAGAGTCCGTGCTGCAGATGATG............................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................ACAGGGACGGTCAGTGACAGCTGCTT................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....TGTGAAGACCATTCGGGATATCATCG...................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTCAGTGTCTGCAGGGCTGAGGAGGACAGGG.................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................ACAGAGAGTCCGTGCTGCAGATGATG............................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GATTGTGAAGACCATTCGGGATATCATCGC..................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCAGTGTCTGCAGGGCTGAGGAGa.......................................................................................................................................................................................... | 25 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGTGAAGACCATTCGGGATATCATCGCC.................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................GATGCAGGCAGGCCAGCGTGTG.......... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCAGTGTCTGCAGGGCTGAGGAG........................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................ACAGAGAGTCCGTGCTGCAGA................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..GATTGTGAAGACCATTCGGGATATCATC....................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCAGTGTCTGCAGGGCTGAGGgg........................................................................................................................................................................................... | 24 | gg | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGGGCATAGGACAGCACTCTAA.......................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTCAGTGTCTGCAGGGCTGAGGAGGg......................................................................................................................................................................................... | 26 | g | 1.00 | 41.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................AGATGATGCAGGCAGGCCAGCGTGT........... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................TGCAGGCAGGCCAGCGTGTGGT........ | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCAGTGTCTGCAGGGCTGAGGAGGgc........................................................................................................................................................................................ | 27 | gc | 1.00 | 41.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................TCATCGCCTTGAACCCTCTGTACAGaga................................................................................................................................................................................................................ | 28 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................................................................GAGAGTCCGTGCTGCAGATGATG............................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCAGTGTCTGCAGGGCTGAGG............................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GAGATTGTGAAGACCATTCGGGATATCATCGCCTTGAACCCTCTGTACAGGTCAGTGTCTGCAGGGCTGAGGAGGACAGGGACGGTCAGTGACAGCTGCTTGACCCCAGTGCACCACACTGCACACTGCCAGAGCTATCTATCACTGAAGCCAGAGTATGTGTAAGGGGCATAGGACAGCACTCTAAGGCACTGGCACTGAACCCCTACAGAGAGTCCGTGCTGCAGATGATGCAGGCAGGCCAGCGTGTGGTGGACAACC ..................................................(((..(((((.((....))....)))))..)))..(((((((.((..((((.......(((((......))))).......))))...)).))))))).((((((((.(((((.....))))).......)))))..)))....................................................................... ..................................................51..........................................................................................................................................191.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................GCCAGAGCTATCTATCACT................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GCTATCTATCACTGAAGCCAGAGTATGT.................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................GCCAGAGCTATCTATCACTGAAGCCA............................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ACTGAAGCCAGAGTATGTG................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................CAGGGACGGTCAGTGACAGCTGCTTG............................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CAGAGCTATCTATCACTGAAGCCAGAGTA....................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |