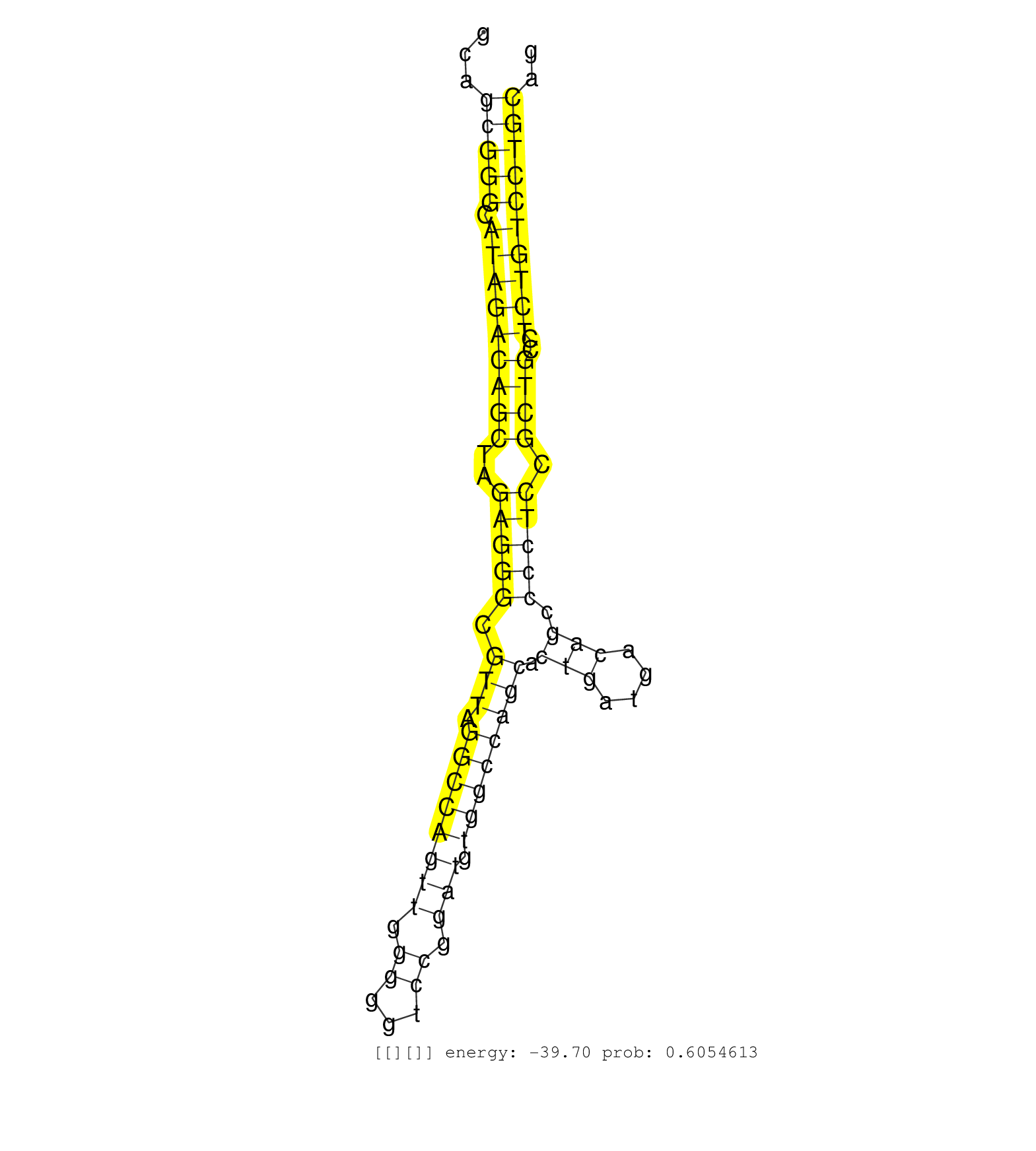
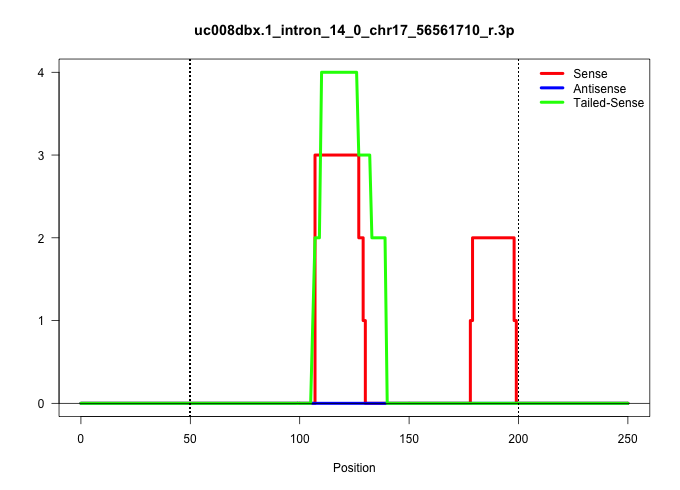
| Gene: Ptprs | ID: uc008dbx.1_intron_14_0_chr17_56561710_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) PIWI.ip |
(10) TESTES |
| GGAAAACACCACCTTAGCTGGGGCTGAGAAGGATCAGCAGGAGCTCAACAGAAAAGGTTTCCCATCCCCCATGTGCCCCAGGAGAGAAGCCTGGGAGCGGCCCCGGCAGCGGGCATAGACAGCTAGAGGGCGTTAGGCCAGTTGGGGGTCCGGATGTGGCCAGCACTGATGACAGCCCCTCCGCTGCCTCTGTCCTGCAGACATTTGCAGCCAGTCCCTTCTCAGACCCCTTCCAGCTGGACAACCCGGA ............................................................................................................(((((.(((((((((..(((((.(((.((((((((.((...)).))).)))))))).(((....))).))))).))))..)))))))))).................................................... .........................................................................................................106...........................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................GGGCATAGACAGCTAGAGGGCGTTAGGCCt.............................................................................................................. | 30 | t | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTCCGCTGCCTCTGTCCTGCAGA................................................. | 23 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| ...............AGCTGGGGCTGAGAAGGATCAGCAGGAG............................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................CAGCGGGCATAGACAGCTAGt........................................................................................................................... | 21 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................GGCGTTAGGCCAGTTt.......................................................................................................... | 16 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................TCCGCTGCCTCTGTCCTGC.................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AGCGGGCATAGACAGCTAGAGGG........................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................AGCGGGCATAGACAGCTAGAGGGtt...................................................................................................................... | 25 | tt | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................AGCGGGCATAGACAGCTAGA........................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................AGCGGGCATAGACAGCTAGAGGGtat..................................................................................................................... | 26 | tat | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................AGCGGGCATAGACAGCTAGAGG......................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTCCGCTGCCTCTGTCCTGCA................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| GGAAAACACCACCTTAGCTGGGGCTGAGAAGGATCAGCAGGAGCTCAACAGAAAAGGTTTCCCATCCCCCATGTGCCCCAGGAGAGAAGCCTGGGAGCGGCCCCGGCAGCGGGCATAGACAGCTAGAGGGCGTTAGGCCAGTTGGGGGTCCGGATGTGGCCAGCACTGATGACAGCCCCTCCGCTGCCTCTGTCCTGCAGACATTTGCAGCCAGTCCCTTCTCAGACCCCTTCCAGCTGGACAACCCGGA ............................................................................................................(((((.(((((((((..(((((.(((.((((((((.((...)).))).)))))))).(((....))).))))).))))..)))))))))).................................................... .........................................................................................................106...........................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............AGCTGGGGCTGAGAAccc............................................................................................................................................................................................................................ | 18 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |