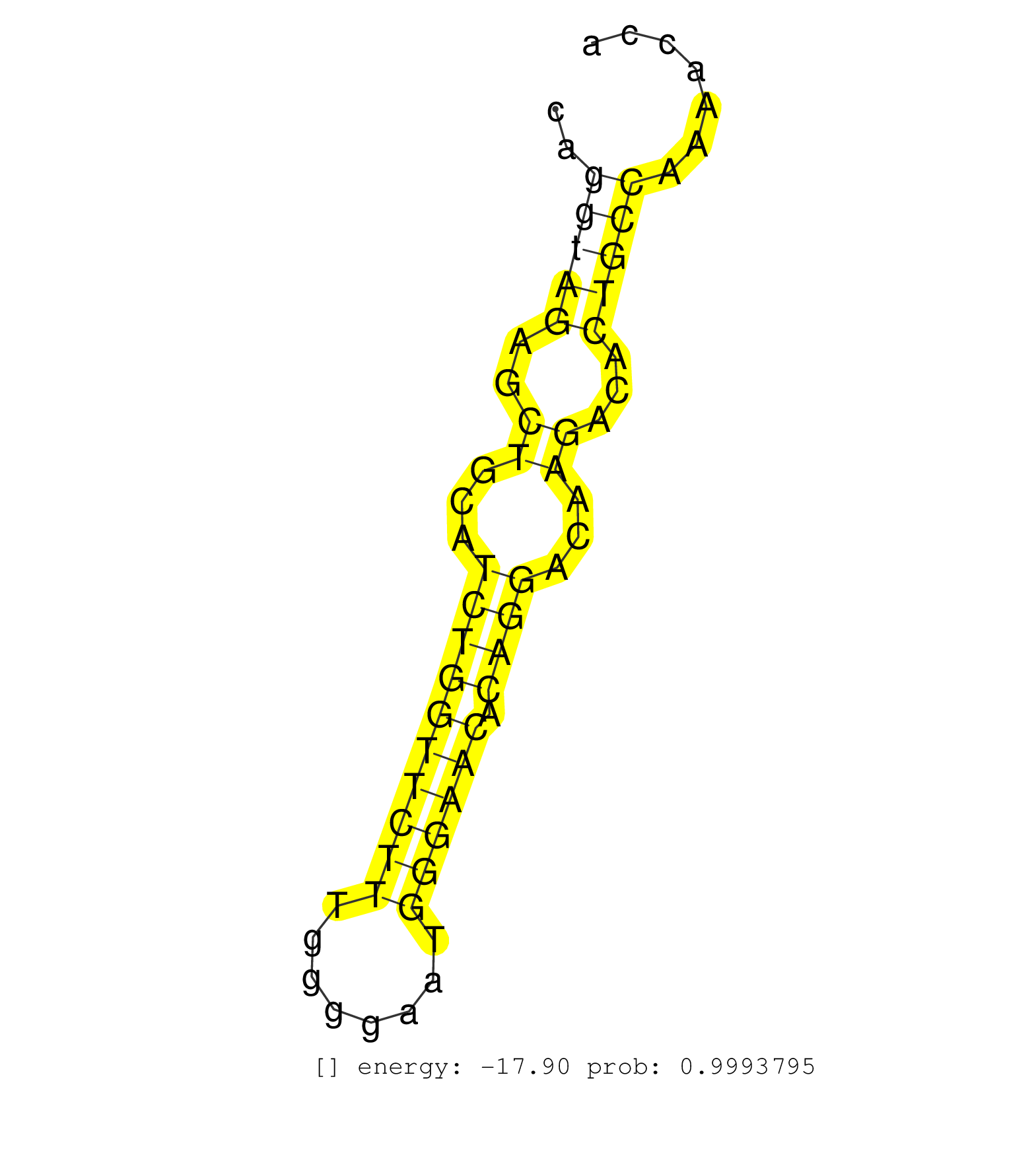

| Gene: Supt3h | ID: uc008cqg.1_intron_9_0_chr17_45178256_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| CACACCTCCTCCTACACCAACAGCTCCCTCTTCTGGATCACAGCATTCAGGTAGAGCTGCATCTGGTTCTTTGGGGAATGGGAACACAGGACAAGACACTGCCAAAACCAAGCAGCGGAAGAGGAAAAAGGTCAGTGCCTTTCTCTTTATCTGTCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTCCTCTTTCTTTCTTTTTTCTTTTCTTCTTCTTCTTTCTTCTTCTTCTTCT .................................................(((((..((...((((((((((........)))))).))))...))...)))))................................................................................................................................................... ...............................................48............................................................110.......................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................TGGGAACACAGGACAAGACACTGCCAAA................................................................................................................................................ | 28 | 1 | 26.00 | 26.00 | 6.00 | 8.00 | 4.00 | - | 3.00 | 2.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................TGGGAACACAGGACAAGACACTGCCAA................................................................................................................................................. | 27 | 1 | 20.00 | 20.00 | 9.00 | 2.00 | 4.00 | - | 3.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................GCATCTGGTTCTTTGGGGAATGGGAACACAGGACAAGACACTGCCAAAACCA............................................................................................................................................ | 52 | 1 | 9.00 | 9.00 | - | - | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGGGAACACAGGACAAGACACTGCCA.................................................................................................................................................. | 26 | 1 | 9.00 | 9.00 | 2.00 | 1.00 | - | - | 1.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................TGGGAACACAGGACAAGACACTGCCAAAAC.............................................................................................................................................. | 30 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................GGACAAGACACTGCCAAAACCAAGCAGCGG.................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| ..............................................................................TGGGAACACAGGACAAGACACTGCCAAAA............................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................GGGAATGGGAACACAGGACAAGACACTGC.................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AACCAAGCAGCGGAAGAGGAAAAAGagca.................................................................................................................... | 29 | agca | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................GGGAATGGGAACACAGGACAA............................................................................................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................AGGACAAGACACTGCCAAAACCAAGCAGC...................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ......................................................................................CAGGACAAGACACTGCCAAAACCAAGCAGC...................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................GGAACACAGGACAAGACACTGCCAAAAC.............................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGGATCACAGCATTCAGGTAGAGCTGCA............................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TCTGGATCACAGCATTCAGGTAGAGag................................................................................................................................................................................................ | 27 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................GCATTCAGGTAGAGCTGCATCTGGTTC..................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TTCTGGATCACAGCATTCAGGTAGAGCT................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGGGAACACAGGACAAGACACTGCCAAt................................................................................................................................................ | 28 | t | 1.00 | 20.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TTCTGGATCACAGCATTCAGGTAGAGC................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGGATCACAGCATTCAGGTAGAGCTG............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................GCCAAAACCAAGCAGCGGAAGAGGAAAt.......................................................................................................................... | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TTTGGGGAATGGGAACACAGGACAAGAC......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................GCCAAAACCAAGCAGCGGAAGAGGAA............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGGGAACACAGGACAAGACACTGCCAtt................................................................................................................................................ | 28 | tt | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGAGCTGCATCTGGTTCTTT.................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................GAGCTGCATCTGGTTCTTTGGG............................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................TTTGGGGAATGGGAACACAGGACAAG........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGCCAAAACCAAGCAGCGGAAGAGGAAA........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGAATGGGAACACAGGACAAGACACT...................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TGGGGAATGGGAACACAGGACAAGAC......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CCAAAACCAAGCAGCGGAAGAGGAtct.......................................................................................................................... | 27 | tct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACCAAGCAGCGGAAGAGGAAAAAGagc..................................................................................................................... | 27 | agc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TCACAGCATTCAGGTAGAGCTGCATC........................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CCAAAACCAAGCAGCGGAAGAGGAAAAAt........................................................................................................................ | 29 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TGGGGAATGGGAACACAGGACAAGACA........................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TTTGGGGAATGGGAACACAGGACAAGACACTG..................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................AGGACAAGACACTGCCAAAACCAAGCAG....................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| CACACCTCCTCCTACACCAACAGCTCCCTCTTCTGGATCACAGCATTCAGGTAGAGCTGCATCTGGTTCTTTGGGGAATGGGAACACAGGACAAGACACTGCCAAAACCAAGCAGCGGAAGAGGAAAAAGGTCAGTGCCTTTCTCTTTATCTGTCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTTCCTCCTCTTTCTTTCTTTTTTCTTTTCTTCTTCTTCTTTCTTCTTCTTCTTCT .................................................(((((..((...((((((((((........)))))).))))...))...)))))................................................................................................................................................... ...............................................48............................................................110.......................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|