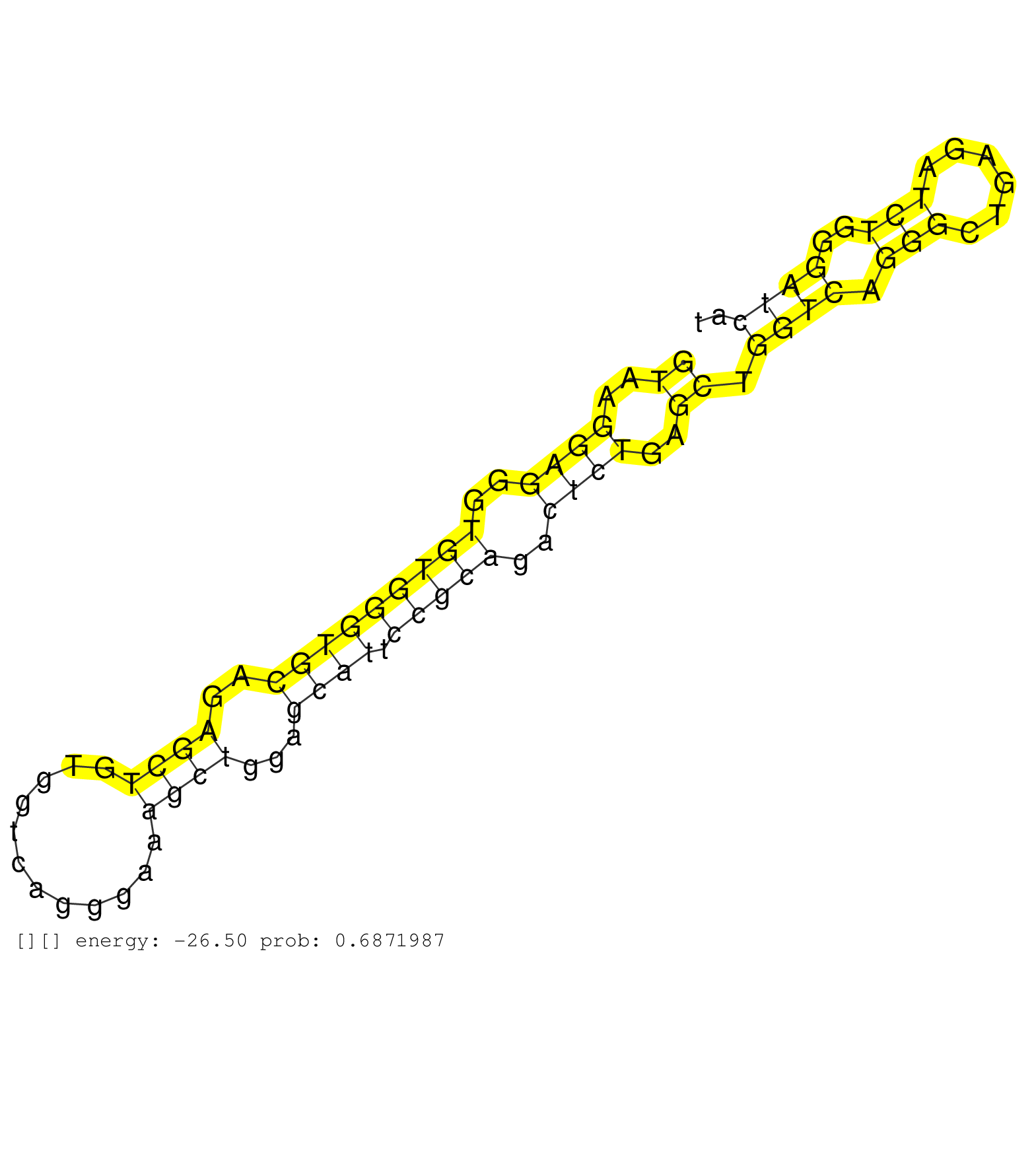

| Gene: H2-Q7 | ID: uc008chs.1_intron_3_0_chr17_35579497_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| GAACCATGAGGGGCTGCCTGAGCCCCTTACCCTGAGATGGGGGAGATGGGGTAAGGAGGGTGTGGGTGCAGAGCTGTGGTCAGGGAAAGCTGGAGCATTCCGCAGACTCTGAGCTGGTCAGGGCTGAGATCTGGGATCATGACCCTCACCTTCATTACCTGTACCTGTCCTCCCAGAGCCTCCTCCATACACTGTCTCCAACATGGCGACCATTGCTGTTGTGGTT ..................................................((..((((..(((((((((..((((............))))...)))).)))))..))))..)).((((.(((......)))..))))........................................................................................ ..................................................51.......................................................................................140.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................TGAGCTGGTCAGGGCTGAGATCTGGGA.......................................................................................... | 27 | 1 | 7.00 | 7.00 | 2.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................TGAGCTGGTCAGGGCTGAGATCTGG............................................................................................ | 25 | 1 | 4.00 | 4.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ................................TGAGATGGGGGAGATGGGGTAAGGAG........................................................................................................................................................................ | 26 | 2 | 3.50 | 3.50 | 1.50 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGAGCTGGTCAGGGCTGAGATCTGGGAT......................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGAGATGGGGGAGATGGGGTAAGGA......................................................................................................................................................................... | 25 | 2 | 3.00 | 3.00 | 2.00 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGAGCTGGTCAGGGCTGAGATCTGGG........................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGAGATGGGGGAGATGGGGTAAGG.......................................................................................................................................................................... | 24 | 2 | 2.00 | 2.00 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ................................TGAGATGGGGGAGATGGGGTAAt........................................................................................................................................................................... | 23 | t | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ................................TGAGATGGGGGAGATGGGGTAAGGAGG....................................................................................................................................................................... | 27 | 2 | 1.50 | 1.50 | 0.50 | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGAGGGTGTGGGTGCAGAGCTGT..................................................................................................................................................... | 27 | 2 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGAGATGGGGGAGATGGGGTAAGGAa........................................................................................................................................................................ | 26 | a | 1.00 | 3.00 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGAGATGGGGGAGATGGGGTAAGta......................................................................................................................................................................... | 25 | ta | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TCATTACCTGTACCTGTCCTCCCAta................................................. | 26 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................TGAGATGGGGGAGATGGGGTAAGGAGt....................................................................................................................................................................... | 27 | t | 1.00 | 3.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................CAGGGAAAGCTGGAGCATTCCGCAGA........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................TTCCGCAGACTCTGAGCTGGTCAGGGC...................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CGCAGACTCTGAGCTGGTCAGGGC...................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................GAAAGCTGGAGCATTCCGCAGACTCTGAGC................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................TCACCTTCATTACCTGTACCTGTCCTCC..................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGGAGCATTCCGCAGACTCTGAGCTGG............................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CCGCAGACTCTGAGCTGGTCAGGGC...................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................ATTCCGCAGACTCTGAGCTGGTCAGG........................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................TGTGGTCAGGGAAAGCTGGAGCATTC.............................................................................................................................. | 26 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGAGCTGTGGTCAGGGAAAGCTGGAG................................................................................................................................... | 26 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................ATGGGGTAAGGAGGGTGTGG................................................................................................................................................................. | 20 | 12 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - |
| ..........................................GAGATGGGGTAAGGAGGGTGT................................................................................................................................................................... | 21 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 |
| GAACCATGAGGGGCTGCCTGAGCCCCTTACCCTGAGATGGGGGAGATGGGGTAAGGAGGGTGTGGGTGCAGAGCTGTGGTCAGGGAAAGCTGGAGCATTCCGCAGACTCTGAGCTGGTCAGGGCTGAGATCTGGGATCATGACCCTCACCTTCATTACCTGTACCTGTCCTCCCAGAGCCTCCTCCATACACTGTCTCCAACATGGCGACCATTGCTGTTGTGGTT ..................................................((..((((..(((((((((..((((............))))...)))).)))))..))))..)).((((.(((......)))..))))........................................................................................ ..................................................51.......................................................................................140.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) |
|---|