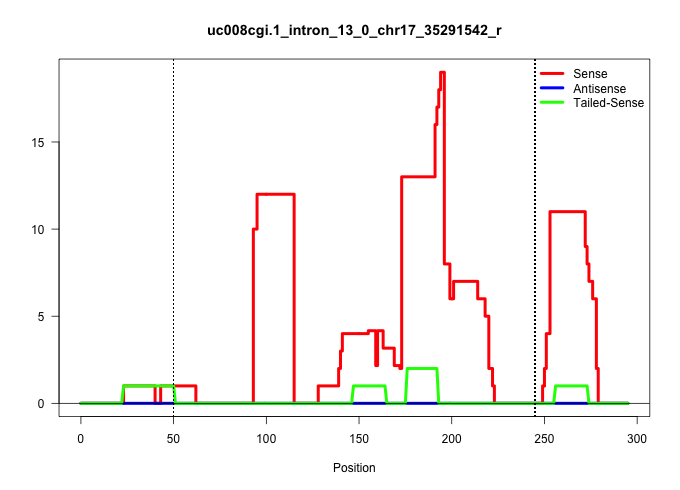
| Gene: Bat2 | ID: uc008cgi.1_intron_13_0_chr17_35291542_r | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(16) TESTES |
| GCCCCCAACCCGTTATGACCCCCCAAGGGCCAGCAGCGCTATCAGTTCTGGTAAGCTGGAGGCAAGCGGGTGGGAATGACTCCATCCCCAAAGGTCAAGTGCTGGAGTGGGGAGCAGGAGTTTGTCATGGGGCCCCTGCTATTGGGATCTGCAGAGCTGGAAGGACTGGGAGGTGGAGTCGGGAGAGCAGGCTAGGGTCAGTGGTAGAATGAGGTAGATGCTGACAGTTTTCCCCTTTTCCCCAGACCCCCACTTCGAGGAGCCAGGGCCGATGGTGAGGGGGGTGGGCGGGACT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................TGGAGTCGGGAGAGCAGGCTAGG................................................................................................... | 23 | 1 | 11.00 | 11.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GTCAAGTGCTGGAGTGGGGAGC.................................................................................................................................................................................... | 22 | 1 | 10.00 | 10.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TTCGAGGAGCCAGGGCCGATGGTGA................. | 25 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................CTAGGGTCAGTGGTAGAATGAGGTAGATG........................................................................... | 29 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CAAGTGCTGGAGTGGGGAGC.................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................ACTTCGAGGAGCCAGGGCCGA....................... | 21 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................AGTCGGGAGAGCAtcag...................................................................................................... | 17 | tcag | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................AAGGACTGGGAGGTGGAGTCGGGAGAGCAGGCTAGGGTC................................................................................................ | 39 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TTCGAGGAGCCAGGGCCGATGGTGAG................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................TTGGGATCTGCAGAGCTG........................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCTGCAGAGCTGGAcgct.................................................................................................................................. | 18 | cgct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................CAAGGGCCAGCAGCGCT............................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................CACTTCGAGGAGCCAGGGCCGATGGT................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................................TGGTAGAATGAGGTAGA............................................................................. | 17 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................GGGTCAGTGGTAGAATGAGGTAGATGCTG........................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................AGTTCTGGTAAGCTGGAGG......................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CAAGGGCCAGCAGCGCTATCAGTTCTGa.................................................................................................................................................................................................................................................... | 28 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................ATTGGGATCTGCAGAGCTGGAAGGACTGG.............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TATTGGGATCTGCAGAGCTGGAAG.................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TTCGAGGAGCCAGGGCCGAT...................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................AGGGTCAGTGGTAGAATGAGGTAGATGCT......................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................GAGGAGCCAGGGCCGATa..................... | 18 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TAGGGTCAGTGGTAGAATGAGG................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................CCACTTCGAGGAGCCAGGGCCGATG..................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................GGGGCCCCTGCTATTGGGATCTGCAGAGCTG........................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GCTGGAAGGACTGGGAG........................................................................................................................... | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| GCCCCCAACCCGTTATGACCCCCCAAGGGCCAGCAGCGCTATCAGTTCTGGTAAGCTGGAGGCAAGCGGGTGGGAATGACTCCATCCCCAAAGGTCAAGTGCTGGAGTGGGGAGCAGGAGTTTGTCATGGGGCCCCTGCTATTGGGATCTGCAGAGCTGGAAGGACTGGGAGGTGGAGTCGGGAGAGCAGGCTAGGGTCAGTGGTAGAATGAGGTAGATGCTGACAGTTTTCCCCTTTTCCCCAGACCCCCACTTCGAGGAGCCAGGGCCGATGGTGAGGGGGGTGGGCGGGACT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|