
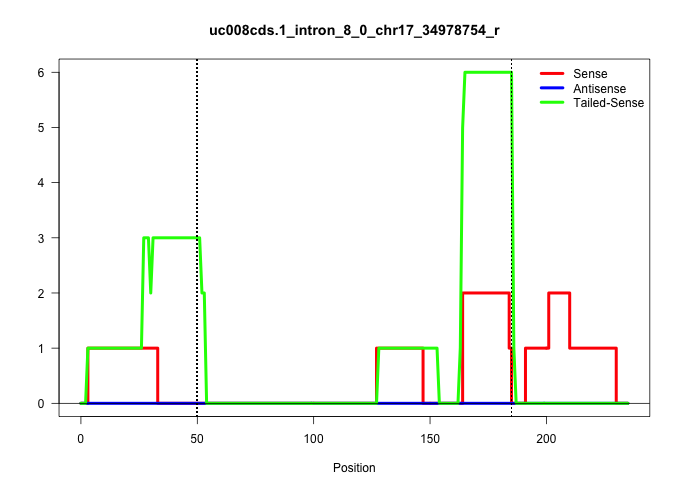
| Gene: Skiv2l | ID: uc008cds.1_intron_8_0_chr17_34978754_r | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(17) TESTES |
| ACATGATGAAGAGGAGCTTCTCAGAGTTCCCATCCCGCAAGGACAGCAAGGTGAGGTTAGGGTGAGCAACGTAGCTGTTGAGACAGGCAGTCTGGGCTGGCCAGGTGGGCATGCGGAGGCAGTGGGGTGGGGGAGGGTGTAGAGGCTGGGCTGGCAGACTCATCACAGCCTCTGCTTCTTCTCAGGCCCATGAGCAGGCTCTGGCTGACCTCACCAAGAGGCTGGGGGCTTTGGA ..................................................................................................................................(((((((.((((((((((((.(((.....))))).)))))))))).))))))).................................................... ...........................................................................................................................124..........................................................185................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................ACAGCCTCTGCTTCTTCTCAGt................................................. | 22 | t | 7.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 |
| ..........................................................................................................................................................................................................................AGGCTGGGGGCTTTGGA | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................GGGGGAGGGTGTAGAGGCTGGGgaat................................................................................. | 26 | gaat | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGGGGGAGGGTGTAGAGGCT........................................................................................ | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCCATCCCGCAAGGACAGCAAGGc....................................................................................................................................................................................... | 25 | c | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGGGGGAGGGTGTAGAGGCTGtag.................................................................................... | 24 | tag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGATGAAGAGGAGCTTCTCAGAGTTCt............................................................................................................................................................................................................. | 27 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGGCTGACCTCACCAAGAGGCTGGGGGCT..... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................ACAGCCTCTGCTTCTTCTCA................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................GAGCAGGCTCTGGCTGACC......................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................TCCCATCCCGCAAGGACAGCAAGGccc..................................................................................................................................................................................... | 27 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................ATCCCGCAAGGACAGCAAGGccc..................................................................................................................................................................................... | 23 | ccc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................ACAGCCTCTGCTTCTTCTCAGat................................................ | 23 | at | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGATGAAGAGGAGCTTCTCAGAGTTCCCAT.......................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................CACAGCCTCTGCTTCTTCTCAGt................................................. | 23 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................................ACAGCCTCTGCTTCTTCTCAG.................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................CAGCCTCTGCTTCTTCTCAGt................................................. | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................CACAGCCTCTGCTTCTTCTCAGa................................................. | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ACATGATGAAGAGGAGCTTCTCAGAGTTCCCATCCCGCAAGGACAGCAAGGTGAGGTTAGGGTGAGCAACGTAGCTGTTGAGACAGGCAGTCTGGGCTGGCCAGGTGGGCATGCGGAGGCAGTGGGGTGGGGGAGGGTGTAGAGGCTGGGCTGGCAGACTCATCACAGCCTCTGCTTCTTCTCAGGCCCATGAGCAGGCTCTGGCTGACCTCACCAAGAGGCTGGGGGCTTTGGA ..................................................................................................................................(((((((.((((((((((((.(((.....))))).)))))))))).))))))).................................................... ...........................................................................................................................124..........................................................185................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|