
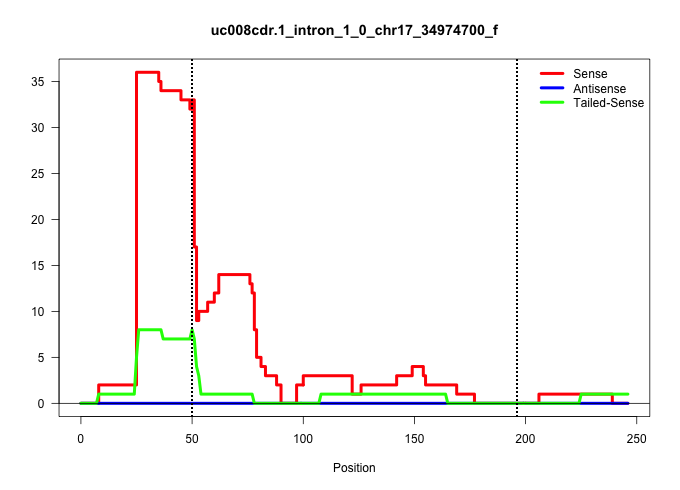
| Gene: Dom3z | ID: uc008cdr.1_intron_1_0_chr17_34974700_f | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(7) PIWI.ip |
(2) PIWI.mut |
(24) TESTES |
| TTGGACCATTTGCTGCGCTGGGTCCTGGAACACCGGAACCAGCTGGAGGGGTGAGCCCGGTGCAACGCAGAAGCTGACCTGGTGACCCGGCTTTCTCTTCTGAAGTTGGGAAAGCCAAATGGTCTTTAGTGGGGACTTTGAGTCAACAGAAGTAAAACTCGAGTCAGACCATAAACCCCGCATTCTTCCCTCCCAGGGGTCCCGGCTGGCTGGCAGGGGCCACGGTGACATGGAGAGGGCACCTGA ......................................................(((.(((.((..((....))))))).)))((((.(((((((((..........)))))))))....)))).......................................................................................................................... ..................................................51.........................................................................126...................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................TGGAACACCGGAACCAGCTGGAGGGG................................................................................................................................................................................................... | 26 | 1 | 46.00 | 46.00 | 27.00 | 8.00 | 5.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................TGGAACACCGGAACCAGCTGGAGGGGT.................................................................................................................................................................................................. | 27 | 1 | 8.00 | 8.00 | - | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................TGGAACACCGGAACCAGCTGGAGGG.................................................................................................................................................................................................... | 25 | 1 | 6.00 | 6.00 | - | 3.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCCGGTGCAACGCAGAAGCTGAC........................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................TGGAACACCGGAACCAGCTGGAGGGGg.................................................................................................................................................................................................. | 27 | g | 2.00 | 46.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGGAACACCGGAACCAGCTGGAGGGGTtt................................................................................................................................................................................................ | 29 | tt | 2.00 | 8.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GGAACACCGGAACCAGCTGGAGGGGg.................................................................................................................................................................................................. | 26 | g | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTGAGCCCGGTGCAACGCAGAAGCTGACC....................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CAACGCAGAAGCTGACCTGGTGACCCGG............................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TTCTGAAGTTGGGAAAGCCAAATGG............................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGGAACACCGGAACCAGCTGGAGG..................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TAGTGGGGACTTTGAGTCAACAGAAGTA............................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGACATGGAGAGGGCACCTGAcaat | 25 | caat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGAGCCCGGTGCAACGCAGAAGCTGA......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................GGAAAGCCAAATGGTCTTTAGTGtgt................................................................................................................ | 26 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................TGAGCCCGGTGCAACGCAGAAGCTGACC....................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................TGGAACACCGGAACCAGCTG......................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGCAACGCAGAAGCTGACCTGGTGACCC.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TGAAGTTGGGAAAGCCAAATGGTCTT........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGGCTGGCAGGGGCCACGGTGACATGGAGAGGG....... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGCCCGGTGCAACGCAGAAGCTGAC........................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TTTGCTGCGCTGGGTCCTGGAACACCG................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCCGGTGCAACGCAGAAGCTG.......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGGAACACCGGAACCAGCTGGAGGGt................................................................................................................................................................................................... | 26 | t | 1.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TTTGCTGCGCTGGGTCCTGGAACACCGGt................................................................................................................................................................................................................. | 29 | t | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TAAACCCCGCATTCTTCCCTCCCAGG................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........TTTGCTGCGCTGGGTCCTGGAACACCGG.................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGCCCGGTGCAACGCAGAAGCTGACCTG..................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCCGGTGCAACGCAGAAGCagac........................................................................................................................................................................ | 28 | agac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCAACAGAAGTAAAACTCGAGTCAGAC............................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTTTGAGTCAACAGAAGTAAAACTCGAGTt................................................................................. | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GGAACACCGGAACCAGCTGGAGGGGgg................................................................................................................................................................................................. | 27 | gg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................AAGTAAAACTCGAGTCAGACCATAAACC..................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGGAACACCGGAACCAGCTGGAtggg................................................................................................................................................................................................... | 26 | tggg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CGGTGCAACGCAGAAGCTGACCTGGT................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TAGTGGGGACTTTGAGTCAACAGAAGTAA........................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTGGACCATTTGCTGCGCTGGGTCCTGGAACACCGGAACCAGCTGGAGGGGTGAGCCCGGTGCAACGCAGAAGCTGACCTGGTGACCCGGCTTTCTCTTCTGAAGTTGGGAAAGCCAAATGGTCTTTAGTGGGGACTTTGAGTCAACAGAAGTAAAACTCGAGTCAGACCATAAACCCCGCATTCTTCCCTCCCAGGGGTCCCGGCTGGCTGGCAGGGGCCACGGTGACATGGAGAGGGCACCTGA ......................................................(((.(((.((..((....))))))).)))((((.(((((((((..........)))))))))....)))).......................................................................................................................... ..................................................51.........................................................................126...................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|