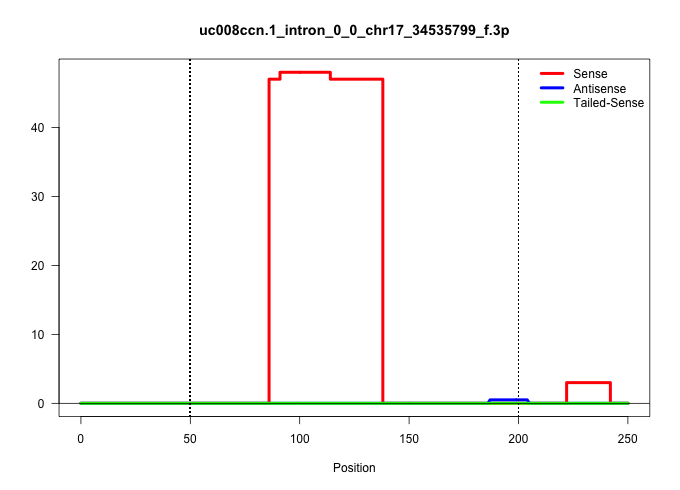
| Gene: BC051142 | ID: uc008ccn.1_intron_0_0_chr17_34535799_f.3p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(1) PIWI.mut |
(4) TESTES |
| AACAAAGTTTTTAAAATTACTATTGAAATTTTAGAGATTTAAATTTATAGATATAAAACTTTCTTTCAAACTACCTCATGAACTTACACTATAAGAATGGAGGAAGGATTGTAACTATCAAGGTTGACTTAAAACAAAGTAGTACTGCCCAGTCCTAAAGGTGGGATATTTGACACTAAAGGGCATCCTTTCTCTTCCAGGGCTGGCTGCTTACCCCAGGAAACAAATCCTCAGACAGTAAGCCCTCAAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|
| ......................................................................................CACTATAAGAATGGAGGAAGGATTGTAACTATCAAGGTTGACTTAAAACAAA................................................................................................................ | 52 | 1 | 47.00 | 47.00 | 47.00 | - | - | - |
| ..............................................................................................................................................................................................................................ACAAATCCTCAGACAGTAAG........ | 20 | 1 | 3.00 | 3.00 | - | 3.00 | - | - |
| ...........................................................................................TAAGAATGGAGGAAGGATTGTAA........................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| AACAAAGTTTTTAAAATTACTATTGAAATTTTAGAGATTTAAATTTATAGATATAAAACTTTCTTTCAAACTACCTCATGAACTTACACTATAAGAATGGAGGAAGGATTGTAACTATCAAGGTTGACTTAAAACAAAGTAGTACTGCCCAGTCCTAAAGGTGGGATATTTGACACTAAAGGGCATCCTTTCTCTTCCAGGGCTGGCTGCTTACCCCAGGAAACAAATCCTCAGACAGTAAGCCCTCAAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................CTTTCTCTTCCAGGGCTG............................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | 0.50 |