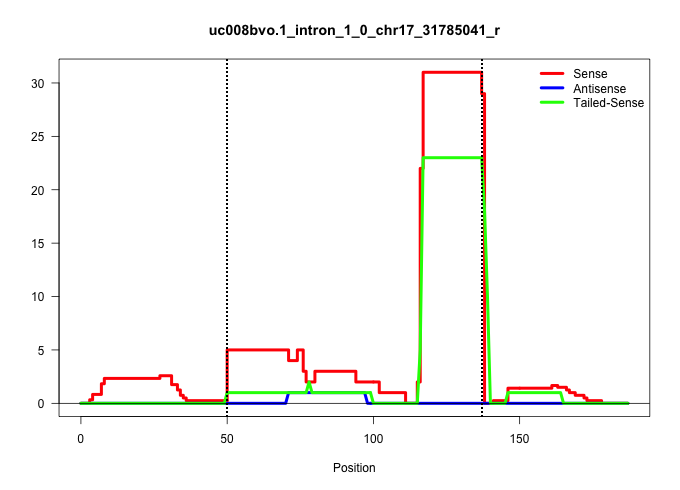
| Gene: U2af1 | ID: uc008bvo.1_intron_1_0_chr17_31785041_r | SPECIES: mm9 |
 |
|
(7) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(24) TESTES |
| TCCCCAGTAACTGACTTCAGGGAAGCCTGCTGCCGCCAGTATGAAATGGGGTAAGGAAGGAAGAGCGGGACTCTGGTGTGGTGGGCAGAGCTCAGTCAGTGGGGGCACAATATGACATGTCTGGATCTGACCCACAGAGAGTGCACAAGAGGGGGCTTCTGCAACTTCATGCATTTGAAGCCCATCT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................ATGTCTGGATCTGACCCACAGA................................................. | 22 | 1 | 19.00 | 19.00 | 5.00 | - | 6.00 | 2.00 | 1.00 | 2.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGA................................................. | 21 | 1 | 9.00 | 9.00 | 4.00 | - | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGAaa............................................... | 23 | aa | 5.00 | 9.00 | - | 3.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGta................................................ | 22 | ta | 5.00 | 0.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................ATGTCTGGATCTGACCCACAGt................................................. | 22 | t | 3.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGt................................................. | 21 | t | 2.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGAAGGAAGAGCGGGACTCTGG............................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGtag............................................... | 23 | tag | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGAAGGAAGAGCGGGAC.................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................TGGTGGGCAGAGCTCAGTCAGTGG..................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................ATGTCTGGATCTGACCCACAGta................................................ | 23 | ta | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................GTGGGCAGAGCTCAGTCAGTGGGGGCACAAT............................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................ATGTCTGGATCTGACCCACAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAGGGGGCTTCTGCAAt...................... | 19 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGAat............................................... | 23 | at | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGAAGGAAGAGCGGGACTCTGGT.............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CATGTCTGGATCTGACCCACAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................ATGTCTGGATCTGACCCACAGtt................................................ | 23 | tt | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGttt............................................... | 23 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................GGTGTGGTGGGCAGAGCTCA............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGGTGGGCAGAGCTCAGTCAGa....................................................................................... | 22 | a | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGtt................................................ | 22 | tt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CATGTCTGGATCTGACCCACAGA................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCTGGATCTGACCCACAGAtt............................................... | 23 | tt | 1.00 | 9.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGAAGGAAGAGCGGGACTCTGGTtt............................................................................................................ | 29 | tt | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGAAGGAAGAGCGGGACTCTGGTG............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CCAGTAACTGACTTCAGGGAAGCCTGCT............................................................................................................................................................ | 28 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| ....CAGTAACTGACTTCAGGGAAGCCTGCTGCC......................................................................................................................................................... | 30 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - |
| ....CAGTAACTGACTTCAGGGAAGCCTGCT............................................................................................................................................................ | 27 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TAACTGACTTCAGGGAAGCCTGCTGCCG........................................................................................................................................................ | 28 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - |
| ...........................TGCTGCCGCCAGTATGAAATGGG......................................................................................................................................... | 23 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - |
| .............................................................................................................................................TGCACAAGAGGGGGCTTCTGCAACT..................... | 25 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAGGGGGCTTCTGCAACTTCATGC............... | 26 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - |
| ..................................................................................................................................................AAGAGGGGGCTTCTGCAACTTCATGCA.............. | 27 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - |
| .......TAACTGACTTCAGGGAAGCCTGCTGCCGC....................................................................................................................................................... | 29 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - |
| .................................................................................................................................................................CAACTTCATGCATTTGA......... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| .......TAACTGACTTCAGGGAAGCCTGCTGCC......................................................................................................................................................... | 27 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAGGGGGCTTCTGCAACTT.................... | 21 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TAACTGACTTCAGGGAAGCCTGCTGC.......................................................................................................................................................... | 26 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - |
| ........AACTGACTTCAGGGAAGCCTGCT............................................................................................................................................................ | 23 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| ..................................................................................................................................................AAGAGGGGGCTTCTGCAACTTCA.................. | 23 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........AACTGACTTCAGGGAAGCCTGCTGC.......................................................................................................................................................... | 25 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - |
| ..................................................................................................................................................AAGAGGGGGCTTCTGCA........................ | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCCCAGTAACTGACTTCAGGGAAGCCTGCTGCCGCCAGTATGAAATGGGGTAAGGAAGGAAGAGCGGGACTCTGGTGTGGTGGGCAGAGCTCAGTCAGTGGGGGCACAATATGACATGTCTGGATCTGACCCACAGAGAGTGCACAAGAGGGGGCTTCTGCAACTTCATGCATTTGAAGCCCATCT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................AGAGCTCAGTCAGTtcc....................................................................................... | 17 | tcc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TCTGGTGTGGTGGGCAGAGCTCAGTCA......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |