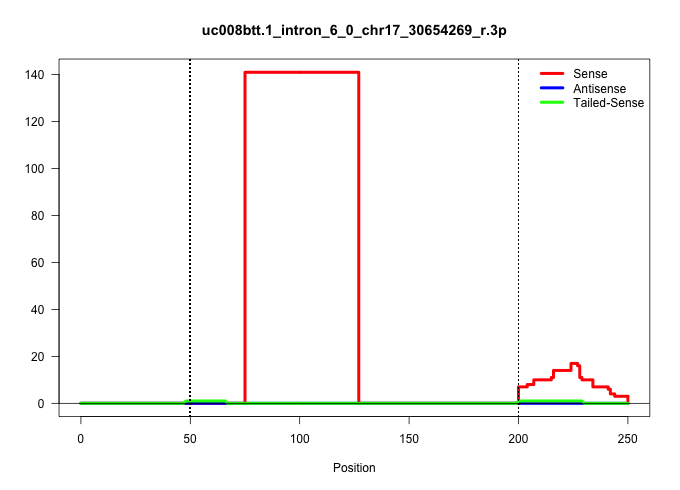
| Gene: Btbd9 | ID: uc008btt.1_intron_6_0_chr17_30654269_r.3p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(11) TESTES |
| TTTAGCTGTGGTAGTGACATTCTGTTTCGAATGCTGAACGCTATCTTAATCTTAGTAGACAGTGTATATTTTCATTTCCATAGACTTGCCTATGCAGCTTAAAATCAGATACAGGTACTGGAATCAGCTAGAATGCTAGAAGCTCTCCTTTCCCTCGTTACCTAGTTACCAAACACTTGCTTTTTCTATTCCAACTGTAGTACCTGAAGAAAACATTGCAACTATGAAGTATGGGGCTCAGGTTGTGAAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................TTCCATAGACTTGCCTATGCAGCTTAAAATCAGATACAGGTACTGGAATCAG........................................................................................................................... | 52 | 1 | 141.00 | 141.00 | 141.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TACCTGAAGAAAACATTGCAACTATGAA...................... | 28 | 1 | 5.00 | 5.00 | - | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGAAGTATGGGGCTCAGGTTGTGAAA | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................TGCAACTATGAAGTATGGGGCTCAGG........ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................................AGAAAACATTGCAACTATGAAGTATGG................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TACCTGAAGAAAACATTGCAACTATGAAG..................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGAAGAAAACATTGCAACTATGAAGTATGG................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................................................................TGCAACTATGAAGTATGGGGCTCAGGTT...... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TACCTGAAGAAAACATTGCAACTATGAAGa.................... | 30 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................ATCTTAGTAGACAGTttga....................................................................................................................................................................................... | 19 | ttga | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................................................TTGCAACTATGAAGTATGGGGCTCAG......... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TACCTGAAGAAAACATTGCAACTATGA....................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| TTTAGCTGTGGTAGTGACATTCTGTTTCGAATGCTGAACGCTATCTTAATCTTAGTAGACAGTGTATATTTTCATTTCCATAGACTTGCCTATGCAGCTTAAAATCAGATACAGGTACTGGAATCAGCTAGAATGCTAGAAGCTCTCCTTTCCCTCGTTACCTAGTTACCAAACACTTGCTTTTTCTATTCCAACTGTAGTACCTGAAGAAAACATTGCAACTATGAAGTATGGGGCTCAGGTTGTGAAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TTCTATTCCAACTGTttta.................................................... | 19 | ttta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |