
| Gene: SRPK1 | ID: uc008brh.1_intron_9_0_chr17_28742332_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(13) TESTES |
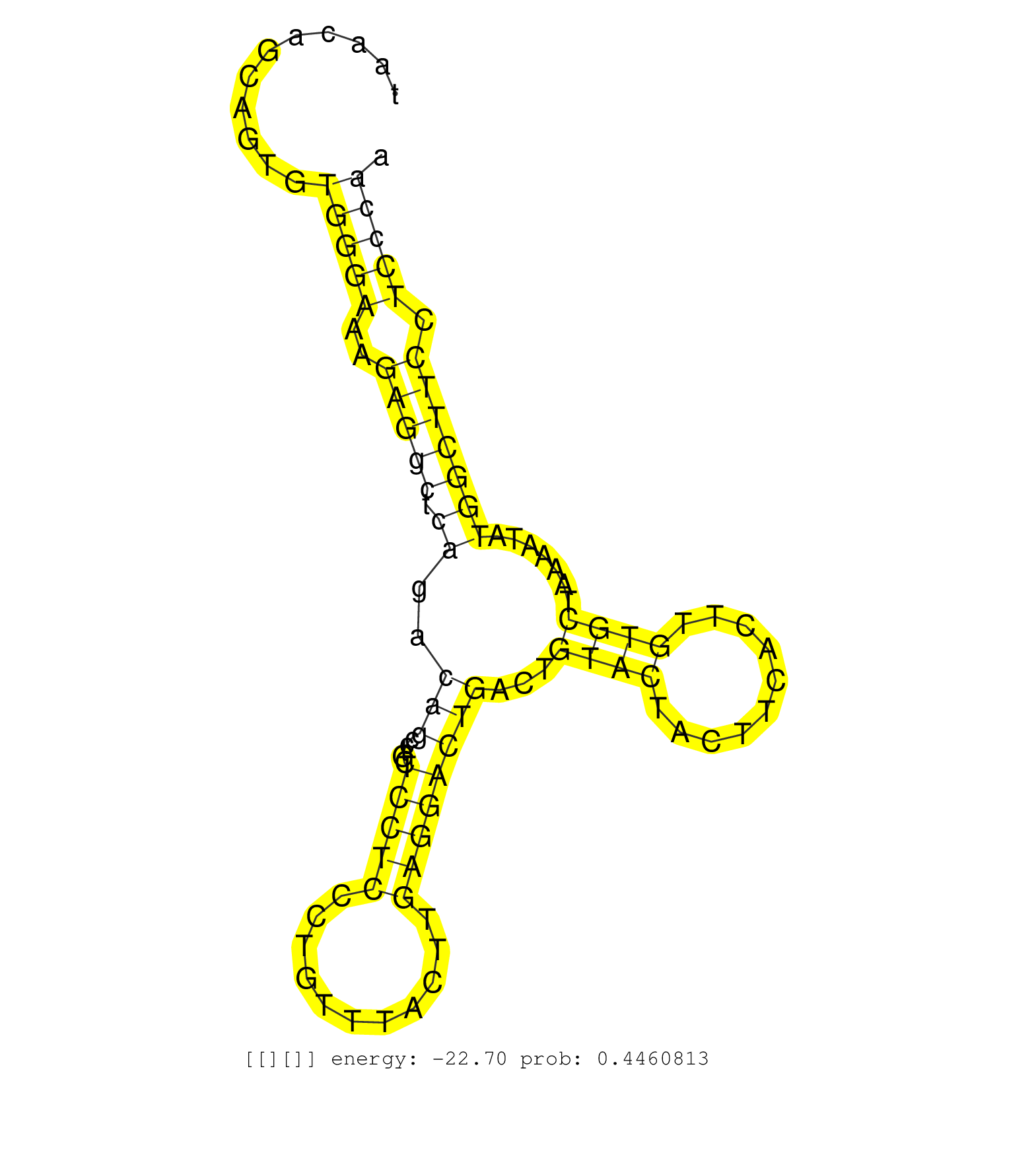
| CTAATTTTGCTGCTATTAGGAGTCATAAATATACCTCAACACAAGTTTATCTTACCCAAAGCCTTTCTTTTCATTTTTTATGGTGGGTTGTAGTAACAGCAGTGTGGGAAAGAGGCTCAGACAGCCTGCTCCTCCCTGTTTACTTGAGGACTGACTGTACTACTTCACTTGTGCTAAAATATGGCTTCCTCCCAACACAGGTTCGCAACTCAGACCCGAATGATCCAAATGGAGAAATGGTTGTTCAACT ........................................................................................................(((((..(((((.((..(((.....(((((...........))))))))...((((..........)))).......))))))).)))))........................................................ .............................................................................................94...................................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................GCTCCTCCCTGTTTACTTGAGGACTGACTGTACTACTTCACTTGTGCTAAAA....................................................................... | 52 | 1 | 79.00 | 79.00 | 79.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGATCCAAATGGAGAAATGGTTGTTCAAC. | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................................................................................GATCCAAATGGAGAAATGGTTGTTC.... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| ............................................................................................................................................................................................................................TGATCCAAATGGAGAAATGGTTGTTC.... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGATCCAAATGGAGAAATGGTTGgtc.... | 26 | gtc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................TTGAGGACTGACTGTcaag........................................................................................ | 19 | caag | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGATCCAAATGGAGAAATGGTTGTT..... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGATCCAAATGGAGAAATGGTTGTTCt... | 27 | t | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGATCCAAATGGAGAAATGGTTGTTCA... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CGCAACTCAGACCCGAATGATCCAtaa.................... | 27 | taa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GAATGATCCAAATGGAGAAATGGTTG....... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGATCCAAATGGAGAAATGGTTGTTCAACT | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................................................................................................GGAGAAATGGTTGTTCA... | 17 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| CTAATTTTGCTGCTATTAGGAGTCATAAATATACCTCAACACAAGTTTATCTTACCCAAAGCCTTTCTTTTCATTTTTTATGGTGGGTTGTAGTAACAGCAGTGTGGGAAAGAGGCTCAGACAGCCTGCTCCTCCCTGTTTACTTGAGGACTGACTGTACTACTTCACTTGTGCTAAAATATGGCTTCCTCCCAACACAGGTTCGCAACTCAGACCCGAATGATCCAAATGGAGAAATGGTTGTTCAACT ........................................................................................................(((((..(((((.((..(((.....(((((...........))))))))...((((..........)))).......))))))).)))))........................................................ .............................................................................................94...................................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................CCGAATGATCCActc....................... | 15 | ctc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGGACTGACTGTACTAtc........................................................................................ | 18 | tc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................GAAAGAGGCTCAGACagcc................................................................................................................................ | 19 | agcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |