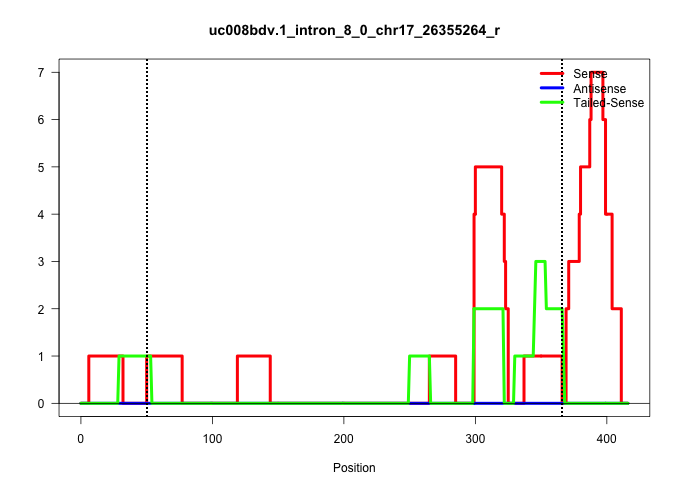
| Gene: Itfg3 | ID: uc008bdv.1_intron_8_0_chr17_26355264_r | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| CAAAAATACCAACAGCAGTAACAATCTCACCAGATCCTGTGCTGATGAAGGCAAGTCTTGTTCTGTCTACTGCGGTGGCCACATGGGAAGAGGGCTCTTTGTCCTGGCTCCCCTTCTCAGTTAGCTTTGGATGATGGGGATTGAGGGAACCACTTCATCATATTTTCCTCTTCATAGATTAGACGAGGTAATACTGTTAGCTCCCTTACTCTACCTGGGGCCACCACCAGAGCCCCATGAGGACCAGGGATACGGGGATAATAGAGGAAACGGGCTGTCATGTGCCTGAGACACAGAATAAGGAAGACAGGAGGTGGCATGCGATAGTTTGAGACTGACCAAGTGGCTGCCACCTCTACCTTCCAGGCTTTTCCACTCCTTGTGCCTTTGTGGTTGCGGTGTCAGGAGCCAATGGC ..............................................................................................................................................................................................................................................................................(((((........))))).............(((((...((((((((((.((.((.(((.......)))...)).))))))))))))..))))).................................................... ..........................................................................................................................................................................................................................................................................267................................................................................................366................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................................................................................................................................................CTGCCACCTCTACCTTCCAGt................................................. | 21 | t | 6.00 | 0.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................GCTGCCACCTCTACCTTCCAGa................................................. | 22 | a | 3.00 | 0.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................TTTCCACTCCTTGTGCCTTTGTGGTTGCGG................. | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................AAGGAAGACAGGAGGTGGCAT................................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................................................................................................................AAGGAAGACAGGAGGTGGCATGCGAT........................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................................................................TGTGGTTGCGGTGTCAGGAGCCA..... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................TGTGCCTTTGTGGTTGCGGTGTCA............ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTCTTGTTCTGTCTACTGCGGTG................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................CTGCCACCTCTACCTTCCAGtt................................................ | 22 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....ATACCAACAGCAGTAACAATCT..................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................AAGGAAGACAGGAGGTGGCATGCG............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................TTGTGCCTTTGTGGTTGCGGTGTCA............ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................GAGACTGACCAAGTGGCTGCCACa.............................................................. | 24 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TACCAACAGCAGTAACAATCTCACCA................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................................................................................................................................................................................................AAGGAAGACAGGAGGTGGCATtc.............................................................................................. | 23 | tc | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................CCAGATCCTGTGCTGATGAAGGCtt.......................................................................................................................................................................................................................................................................................................................................................................... | 25 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................AAGGAAGACAGGAGGTGGCATGC.............................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................GTTAGCTTTGGATGATGGGGATTGA................................................................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................AAGGAAGACAGGAGGTGGCATGt.............................................................................................. | 23 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................GCTGCCACCTCTACCTTCCAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................AGGAAGACAGGAGGTGGCATGCG............................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................TTGTGGTTGCGGTGTCAGGAGCCA..... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................TCCACTCCTTGTGCCTTTGTGGTTGC................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................ACCAAGTGGCTGCCACCTCTACCTTCCAG.................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................................................................................TACGGGGATAATAtga...................................................................................................................................................... | 16 | tga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................GGAAACGGGCTGTCATGTGC................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAAAAATACCAACAGCAGTAACAATCTCACCAGATCCTGTGCTGATGAAGGCAAGTCTTGTTCTGTCTACTGCGGTGGCCACATGGGAAGAGGGCTCTTTGTCCTGGCTCCCCTTCTCAGTTAGCTTTGGATGATGGGGATTGAGGGAACCACTTCATCATATTTTCCTCTTCATAGATTAGACGAGGTAATACTGTTAGCTCCCTTACTCTACCTGGGGCCACCACCAGAGCCCCATGAGGACCAGGGATACGGGGATAATAGAGGAAACGGGCTGTCATGTGCCTGAGACACAGAATAAGGAAGACAGGAGGTGGCATGCGATAGTTTGAGACTGACCAAGTGGCTGCCACCTCTACCTTCCAGGCTTTTCCACTCCTTGTGCCTTTGTGGTTGCGGTGTCAGGAGCCAATGGC ..............................................................................................................................................................................................................................................................................(((((........))))).............(((((...((((((((((.((.((.(((.......)))...)).))))))))))))..))))).................................................... ..........................................................................................................................................................................................................................................................................267................................................................................................366................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................................................................................................................................................................TGGTTGCGGTGTCAtttc............ | 18 | tttc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |