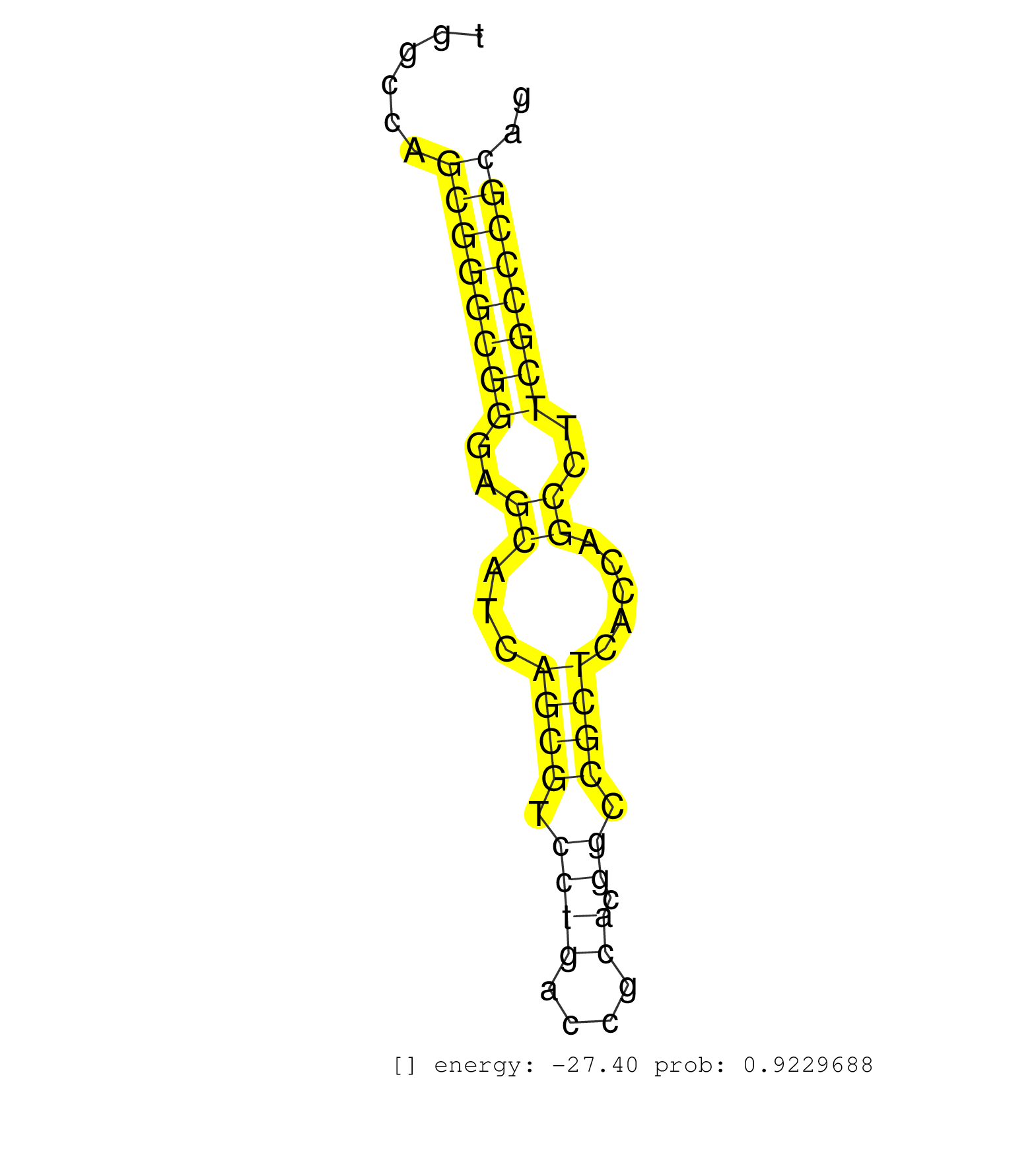

| Gene: Rab40c | ID: uc008bco.1_intron_5_0_chr17_26056495_r | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(1) OVARY |
(2) PIWI.mut |
(22) TESTES |
| CGGCGGGTGGCGGCGGCGGCAGCGGCGGCGGCGGCGGCTAGGAGGCTGAGGTGAAGCTGGCGGCAGCGGGGCGGGTGCACTGGGGGCGGTGGCCAGCGGGCGGGAGCATCAGCGTCCTGACCGCACGGCCGCTCACCAGCCTTCGCCCGCAGGTCTGGCGGCGGTGCCGTGGGCCGCGGTGCGGCGGGCGGGAGGATGGGCA ...............................................................................................((((((((..((...((((.((((....)).)).)))).....))..)))))))).................................................... .........................................................................................90............................................................152................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT4() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................AGCGGGCGGGAGCATCAGCGT....................................................................................... | 21 | 1 | 8.00 | 8.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - |
| .....................................................................................................................................................................................CGGCGGGCGGGAGGAgct... | 18 | gct | 5.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| .........................................GAGGCTGAGGTGAAGCTGGCGGCA......................................................................................................................................... | 24 | 1 | 4.00 | 4.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................AGGAGGCTGAGGTGAAGCTGG.............................................................................................................................................. | 21 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AGCGGGCGGGAGCATCAGCGTC...................................................................................... | 22 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................CTAGGAGGCTGAGGTGAAGCTGGCGGC.......................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCGCTCACCAGCCTTCGCCCG..................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CAGCGGGCGGGAGCATCAGCGT....................................................................................... | 22 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AGCGGGCGGGAGCATCAGCG........................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................CAGCGGGCGGGAGCATCAGCGTt...................................................................................... | 23 | t | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TAGGAGGCTGAGGTGAAGCTGGCGGC.......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CGGCGGGCGGGAGGAgctg.. | 19 | gctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................AGCGGGCGGGAGCATCAGagtc...................................................................................... | 22 | agtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GGGTGGCGGCGGCGGttt.................................................................................................................................................................................... | 18 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GCCCGCAGGTCTGGCGGCGG...................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................AGGCTGAGGTGAAGCTGGCGGCAGCGGG.................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGCGGCGGGCGGGAGGcga.... | 19 | cga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................GGCTAGGAGGCTGAGGTGAAGCTGG.............................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CAGCGGGCGGGAGCATCAGCGTCt..................................................................................... | 24 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CGGCGGCGGCGGCTAGGA............................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AGCGGGCGGGAGCATCAGCGTCt..................................................................................... | 23 | t | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................GTGCACTGGGGGCGGTGGCC............................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CAGCGGGCGGGAGCATCAGC......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GCGGGTGGCGGCGGCGGC...................................................................................................................................................................................... | 18 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| CGGCGGGTGGCGGCGGCGGCAGCGGCGGCGGCGGCGGCTAGGAGGCTGAGGTGAAGCTGGCGGCAGCGGGGCGGGTGCACTGGGGGCGGTGGCCAGCGGGCGGGAGCATCAGCGTCCTGACCGCACGGCCGCTCACCAGCCTTCGCCCGCAGGTCTGGCGGCGGTGCCGTGGGCCGCGGTGCGGCGGGCGGGAGGATGGGCA ...............................................................................................((((((((..((...((((.((((....)).)).)))).....))..)))))))).................................................... .........................................................................................90............................................................152................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT4() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|