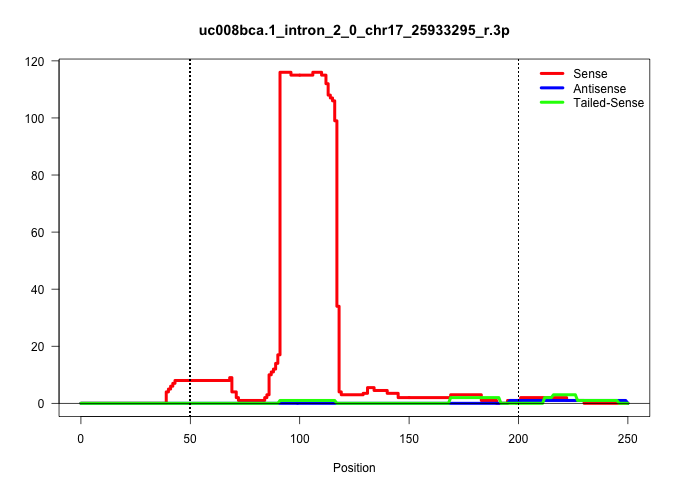
| Gene: Metrn | ID: uc008bca.1_intron_2_0_chr17_25933295_r.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(27) TESTES |
| GCCTGACCCTGGGCGGCCCCGATCCGGGCACACGGCCCAGCATCGTCTGTCTGCGCCCAGAGCGGCCCTTCGCTGGTGCCCAGGTCTTCGCTGAACGTATGACCGGCAATCTAGAGTTGCTACTGGCCGAGGGCCCGGACCTGGCTGGGGGCCGCTGCATGCGCTGGGGTCCCCGCGAGCGCCGAGCCCTTTTCCTGCAGGCCACACCACACCGCGACATCAGCCGCAGAGTTGCTGCCTTCCGTTTTGA ........................................................................................................((((((.....))))))...((((...))))(((..((.((.(((((((.((((....)).))))))))))).))..))).................................................................. ......................................................................................87..................................................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................TGAACGTATGACCGGCAATCTAGAGT..................................................................................................................................... | 26 | 1 | 64.00 | 64.00 | 25.00 | 16.00 | - | 7.00 | 10.00 | 1.00 | - | - | - | 2.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGAACGTATGACCGGCAATCTAGAGTT.................................................................................................................................... | 27 | 1 | 30.00 | 30.00 | 6.00 | 7.00 | - | 7.00 | 3.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................GCATCGTCTGTCTGCGCCCAGAGCGGCCCT..................................................................................................................................................................................... | 30 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGAACGTATGACCGGCAATCTAGAG...................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................TTCGCTGAACGTATGACCGGCAATCT.......................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTCGCTGAACGTATGACCGGCAATCTA......................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CCAGCATCGTCTGTCTGCGCCCAGAGCGGCCCTTCG.................................................................................................................................................................................. | 36 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CTGAACGTATGACCGGCAATCTAGAG...................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GGCCCGGACCTGGCTGGGGGCCGCTGCATGCGCTGGGGTCCCCGCGAGCGCC................................................................... | 52 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TCCCCGCGAGCGCCGAGCCCaat.......................................................... | 23 | aat | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CGCGACATCAGatca....................... | 15 | atca | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TACTGGCCGAGGGCCCGGACCTGGC......................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGAACGTATGACCGGCAATCTAGAGTTG................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CGCTGAACGTATGACCGGCAATCTAGAGT..................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TTCGCTGGTGCCCAGGTCTTCGCTGAAC.......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................GCTGAACGTATGACCGGCAATCTAGAG...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................CCAGGTCTTCGCTGAACGTATGA.................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTCGCTGAACGTATGACCGGCAATC........................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................TAGAGTTGCTACTGGCCGAGGGCCCGGAC.............................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTCGCTGAACGTATGACCGGCAAT............................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCTTCGCTGAACGTATGACCGGCAATCTA......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TCGTCTGTCTGCGCCCAGAGCGGCCCTTC................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GCTGAACGTATGACCGGCAATCTAG........................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGGCCACACCACACCGCGACATCA............................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................CTGAACGTATGACCGGCAATCTAGAGT..................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGAACGTATGACCGGCAATCTAGAGg..................................................................................................................................... | 26 | g | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CTTCGCTGAACGTATGACCGGCAATCTA......................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TCCCCGCGAGCGCCGAGCCCT............................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................TCGCTGAACGTATGACCGGCAATCTAGA....................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................ATCGTCTGTCTGCGCCCAGAGCGGCCCTTC................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................CATCGTCTGTCTGCGCCCAGAGCGGCCCT..................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................CAATCTAGAGTTGCTACTGGCCGAGGGC.................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................CGTCTGTCTGCGCCCAGAGCGGCCCTTCG.................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CGGACCTGGCTGGGagaa................................................................................................. | 18 | agaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................TGAACGTATGACCGGCAATCTAGAGTTGC.................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................ACATCAGCCGCAGAGTTGCTGCCTTCCctt.... | 30 | ctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................CCACACCACACCGCGACATCAGCCGCAGA.................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AGGGCCCGGACCTGGC......................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCCTGACCCTGGGCGGCCCCGATCCGGGCACACGGCCCAGCATCGTCTGTCTGCGCCCAGAGCGGCCCTTCGCTGGTGCCCAGGTCTTCGCTGAACGTATGACCGGCAATCTAGAGTTGCTACTGGCCGAGGGCCCGGACCTGGCTGGGGGCCGCTGCATGCGCTGGGGTCCCCGCGAGCGCCGAGCCCTTTTCCTGCAGGCCACACCACACCGCGACATCAGCCGCAGAGTTGCTGCCTTCCGTTTTGA ........................................................................................................((((((.....))))))...((((...))))(((..((.((.(((((((.((((....)).))))))))))).))..))).................................................................. ......................................................................................87..................................................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................CCACACCGCGACATCagg............................. | 18 | agg | 26.00 | 0.00 | - | - | 11.00 | - | - | - | 7.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CCACACCGCGACATCaggc............................. | 19 | aggc | 15.00 | 0.00 | - | - | 14.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AGCCGCAGAGTTGCTGCCTTCCGTTTT.. | 27 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CCACACCGCGACATCag............................. | 17 | ag | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CCACACCGCGACAagg............................... | 16 | agg | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................CCGCGAGCGCCGAGCCCTTTTCCTGCAa................................................... | 28 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................GTCTGTCTGCGCCCAGAGCG.......................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GCAGGCCACACCACACCGCGACATCA............................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................GCCGCAGAGTTGCTGCCTTCCGTTTTGA | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |