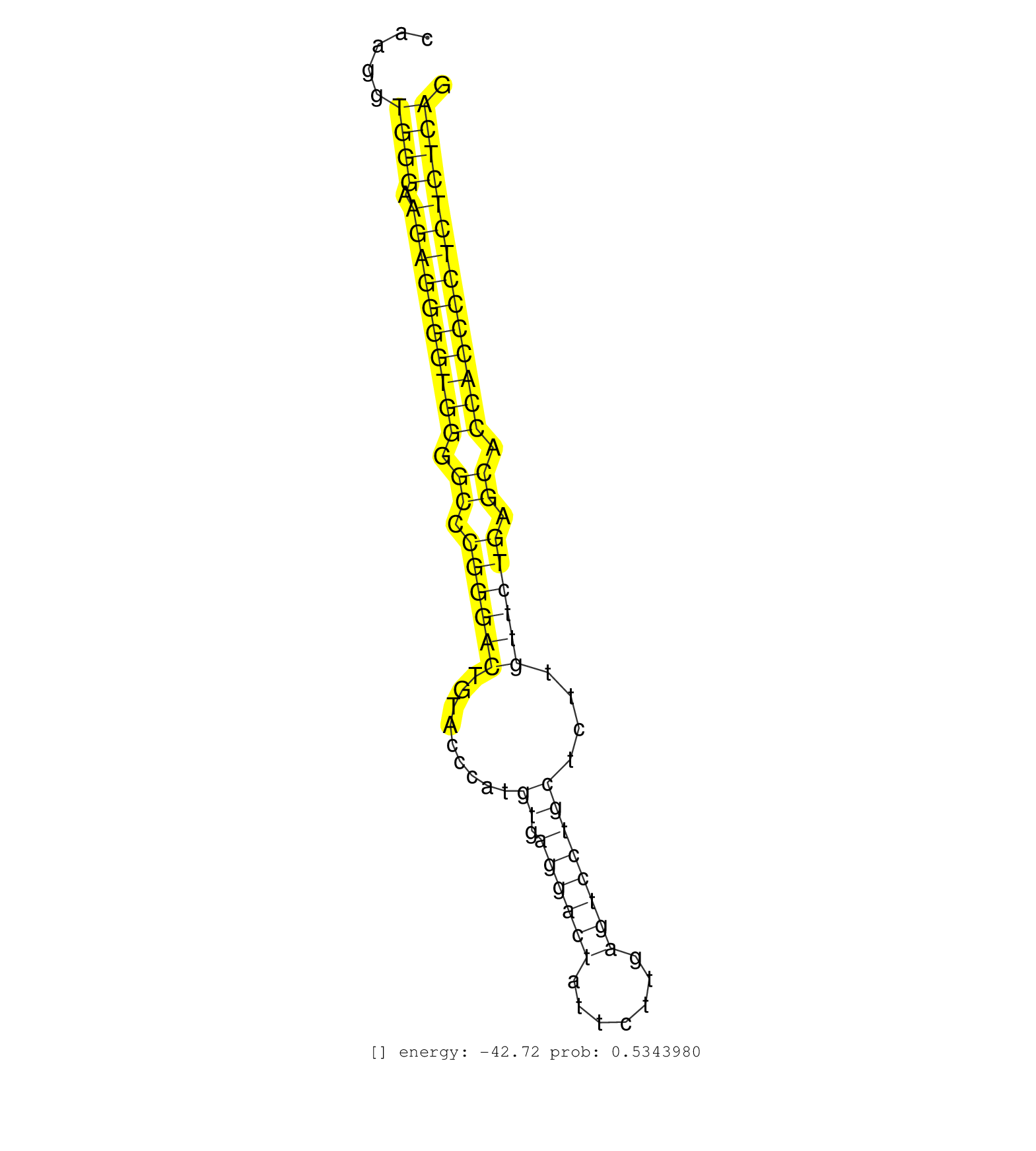
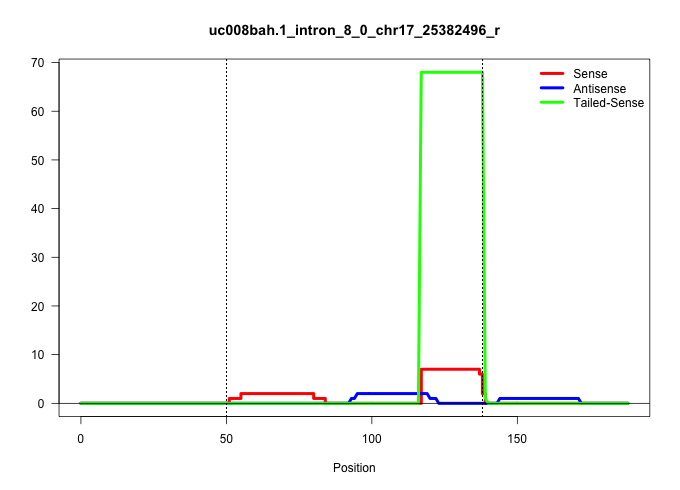
| Gene: Baiap3 | ID: uc008bah.1_intron_8_0_chr17_25382496_r | SPECIES: mm9 |
 |
 |
|
(9) OTHER.mut |
(1) OVARY |
(1) PIWI.ip |
(1) PIWI.mut |
(19) TESTES |
| AAGACCTCCACCGGGAGGCCCACACTGTGACAGCACACCTGACTTCCAAGGTGGGAAGAGGGGTGGGGCCCGGGACTGTACCCATGTGAGGACTATTCTTGAGTCCTGCTCTTGTTCTGAGCACCACCCCTCTCTCAGATGGTGGCCGACATCAGGAAGTACATACAGCACATCAGCCTGTCCCCTGA ...................................................((((.((((((((((.((.((((((.........((.((((((.......))))))))....)))))).)).))))))))))))))................................................... ..............................................47.........................................................................................138................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................TGAGCACCACCCCTCTCTCAGt................................................. | 22 | t | 97.00 | 5.00 | 17.00 | 11.00 | 15.00 | 1.00 | 10.00 | 10.00 | 5.00 | 7.00 | 5.00 | 2.00 | 3.00 | 3.00 | 3.00 | 2.00 | - | 2.00 | - | - | 1.00 | - |
| .....................................................................................................................TGAGCACCACCCCTCTCTCAGA................................................. | 22 | 1 | 16.00 | 16.00 | 1.00 | - | - | 11.00 | - | - | 1.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGAGCACCACCCCTCTCTCAG.................................................. | 21 | 1 | 5.00 | 5.00 | - | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGAGCACCACCCCTCTCTCAGc................................................. | 22 | c | 2.00 | 5.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGAGCACCACCCCTCTCTCA................................................... | 20 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................TGGGAAGAGGGGTGGGGCCCGGGACTGTA............................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................AAGAGGGGTGGGGCCCGGGACTGTACCCA........................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................TGAGCACCACCCCTCTCTCcgt................................................. | 22 | cgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGAGCACCACCCCTCTCTCAGtt................................................ | 23 | tt | 1.00 | 5.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGACCTCCACCGGGAGGCCCACACTGTGACAGCACACCTGACTTCCAAGGTGGGAAGAGGGGTGGGGCCCGGGACTGTACCCATGTGAGGACTATTCTTGAGTCCTGCTCTTGTTCTGAGCACCACCCCTCTCTCAGATGGTGGCCGACATCAGGAAGTACATACAGCACATCAGCCTGTCCCCTGA ...................................................((((.((((((((((.((.((((((.........((.((((((.......))))))))....)))))).)).))))))))))))))................................................... ..............................................47.........................................................................................138................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................GCCGACATCAGGAAGTACATACAGCACA................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TATTCTTGAGTCCTGCTCTTGTTCTGA.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................TTCTTGAGTCCTGCTCTTGTTCTGAGCA................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |