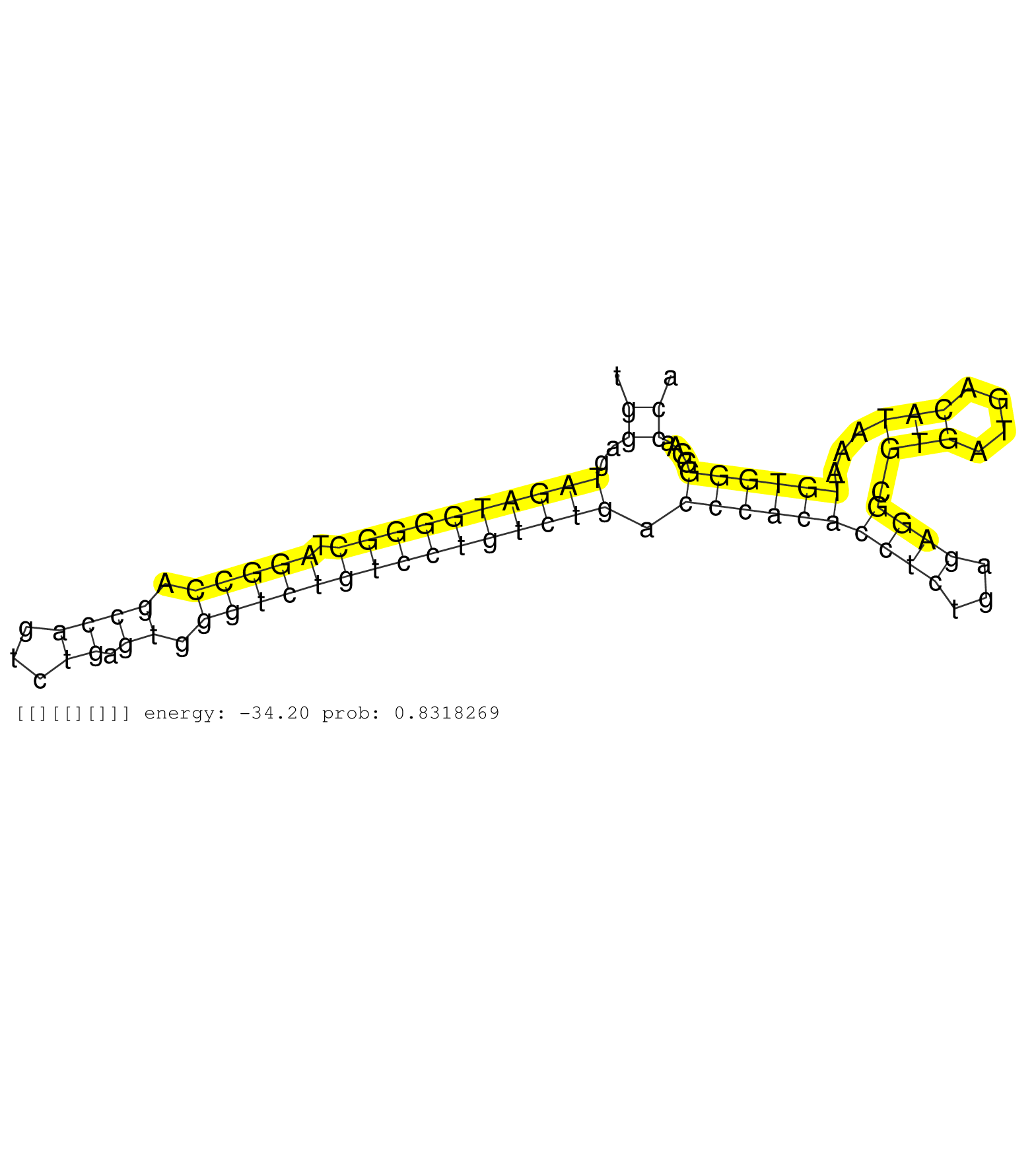

| Gene: Telo2 | ID: uc008azq.1_intron_0_0_chr17_25237066_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| GCTGGGGGACCTGCCAGATGAGCTACTGGAAGCCAGATCCTGGTTGGCAGGTTAGTGTCTGGACCAGAACAGGGACACCAGATGTGGAGTGGCTGGAGTAGATGGGGCTAGGCCAGCCAGTCTGAGTGGGTCTGTCCTGTCTGACCCACACCTCTGAGAGGCGTGATGACATAAATGTGGGGGAACCAGAGGACAACTGTTACCCTGTTTCTTTGGGGATTATTACCTGGGTCTCAACAGACAACTCAGA ..............................................................................................((..((((((((((.(((((.((((...)).)).))))))))))))))).((((((((((...)))).(((....)))...))))))....))............................................................... .............................................................................................94............................................................................................188............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT4() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................CGTGATGACATAAATGTGGGGGAACCAGAGGACAACTGTTACCCTGTTTCTT..................................... | 52 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGGCGTGATGACATAAATGTGGGGGA.................................................................. | 26 | 1 | 6.00 | 6.00 | - | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................AGGCGTGATGACATAAATGTGGGGG................................................................... | 25 | 1 | 4.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - |
| ..............................................................................................................................................................AGGCGTGATGACATAAATGTGGGGGAAC................................................................ | 28 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGGCGTGATGACATAAATGTGGG..................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGAGCTACTGGAAGCCAGATCCTGGT.............................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................GAGGCGTGATGACATAA............................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AGCCAGTCTGAGTGGt........................................................................................................................ | 16 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGGCGTGATGACATAAATGTGGGGGAACC............................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GGCGTGATGACATAAATGTGGGGGAAC................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGGCGTGATGACATAAATGagg...................................................................... | 22 | agg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................................AGGCGTGATGACATAAATGTGGGG.................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................TAGATGGGGCTAGttta....................................................................................................................................... | 17 | ttta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTAGTGTCTGGACCAGAACAGGGACAC............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TAAATGTGGGGGAACCAGAGGACAACT.................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGAGCTACTGGAAGCCAGATCCTGGTT............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GGCGTGATGACATAAATGTGG...................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGGCGTGATGACATAAATGTGG...................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGAGCTACTGGAAGCCAGATCCTGGTTGGC.......................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| GCTGGGGGACCTGCCAGATGAGCTACTGGAAGCCAGATCCTGGTTGGCAGGTTAGTGTCTGGACCAGAACAGGGACACCAGATGTGGAGTGGCTGGAGTAGATGGGGCTAGGCCAGCCAGTCTGAGTGGGTCTGTCCTGTCTGACCCACACCTCTGAGAGGCGTGATGACATAAATGTGGGGGAACCAGAGGACAACTGTTACCCTGTTTCTTTGGGGATTATTACCTGGGTCTCAACAGACAACTCAGA ..............................................................................................((..((((((((((.(((((.((((...)).)).))))))))))))))).((((((((((...)))).(((....)))...))))))....))............................................................... .............................................................................................94............................................................................................188............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT4() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................GGAAGCCAGATCCgaga.................................................................................................................................................................................................................. | 17 | gaga | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GGAAGCCAGATCgaga................................................................................................................................................................................................................... | 16 | gaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |