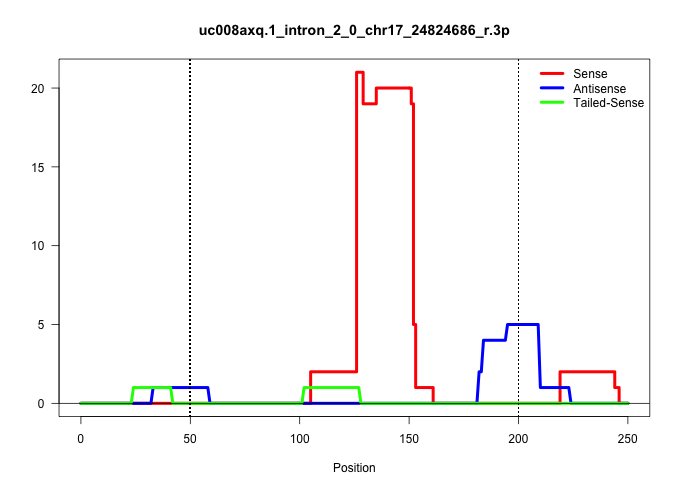
| Gene: Syngr3 | ID: uc008axq.1_intron_2_0_chr17_24824686_r.3p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| CACCACTGCCCGGCATAAGTAGCAGTTTTAATTACTCCTTAAATGCTTCTTTATTTTCAATAATTGTGCAGTACACGAGGGAGTGAGAAATGGGTTCGGTGCTGCAGAGGAACTCTGGGGCAGGTATGGAGACCTGAACTCTTGACAGGGGGAACAGATGCCTCAAGCAGTGGCGTGAGGCTCTGCTTGCTGCTCTGCAGGTGTTTTCTATTGCGGTGTTCGGACCGATTGTCAACGAGGGCTACGTGAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................TGGAGACCTGAACTCTTGACAGGGGG.................................................................................................. | 26 | 1 | 14.00 | 14.00 | 3.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - |
| ..............................................................................................................................TGGAGACCTGAACTCTTGACAGGGGGA................................................................................................. | 27 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - |
| .........................................................................................................AGAGGAACTCTGGGGCAGGTATGG......................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ........................GTTTTAATTACTCCggcg................................................................................................................................................................................................................ | 18 | ggcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................TGGAGACCTGAACTCTTGACAGGGG................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TCGGACCGATTGTCAACGAGGGCTACG.... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TCGGACCGATTGTCAACGAGGGCTA...... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGCAGAGGAACTCTGGGGCAGGTATa.......................................................................................................................... | 26 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GAACTCTTGACAGGGGGAACAGATGC......................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACCACTGCCCGGCATAAGTAGCAGTTTTAATTACTCCTTAAATGCTTCTTTATTTTCAATAATTGTGCAGTACACGAGGGAGTGAGAAATGGGTTCGGTGCTGCAGAGGAACTCTGGGGCAGGTATGGAGACCTGAACTCTTGACAGGGGGAACAGATGCCTCAAGCAGTGGCGTGAGGCTCTGCTTGCTGCTCTGCAGGTGTTTTCTATTGCGGTGTTCGGACCGATTGTCAACGAGGGCTACGTGAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................GCTTGCTGCTCTGCAGGTGTTTTCTA........................................ | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CTGCTTGCTGCTCTGCAGGTGTTTTCTA........................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGGTGTTTTCTATTGCGGTGTTCGGA.......................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................ACTCCTTAAATGCTTCTTTATTTTCA............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |