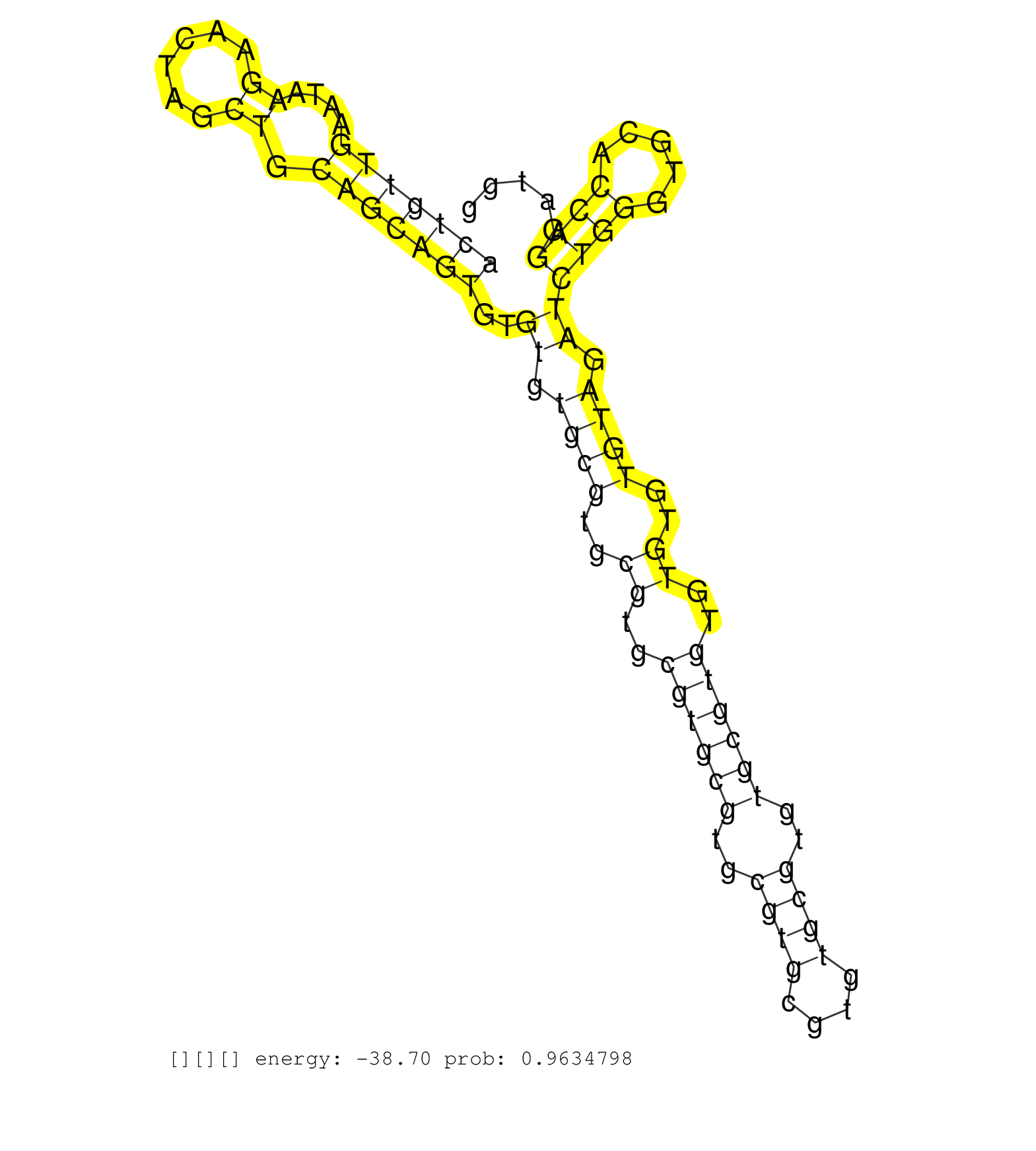
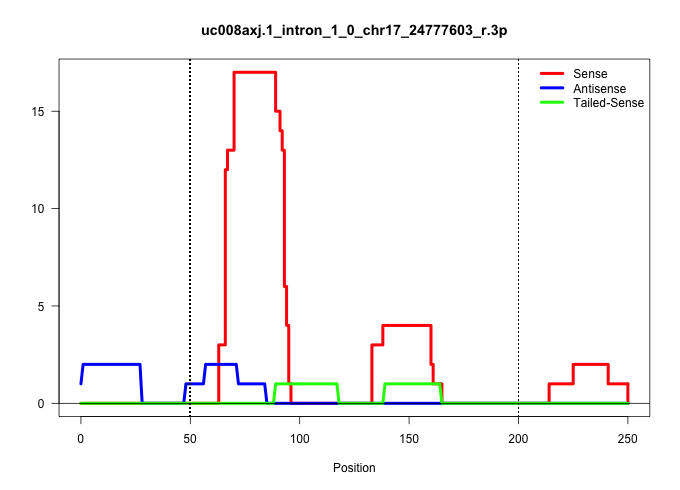
| Gene: Slc9a3r2 | ID: uc008axj.1_intron_1_0_chr17_24777603_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(5) PIWI.ip |
(19) TESTES |
| TTGCCCGTCTGCTGTTAGGTGCCAGCTATGCCCTGGCCCAGACAGTGACCGAAGTGTGCTCACTGTTGAATAAGAACTAGCTGCAGCAGTGTGTGTGCGTGCGTGCGTGCGTGCGTGCGTGTGCGTGTGCGTGTGTGTGTGTAGATCTGGGTGCACCAGGATGGGGATGCTGAAAGCACAACCTATCCTTGGTTTCCTAGCTCAATGGTGGCTCTGTGTGCTCATCCCGAAGTGACCTGCCAGGCTCAGA .............................................................(((((((....((......)).)))))))..((.((((..((..((((((..((((....))))..))))))..))..)))).))((((.....))))........................................................................................... .............................................................62....................................................................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................TGAATAAGAACTAGCTGCAGCAGTGTG............................................................................................................................................................. | 27 | 1 | 7.00 | 7.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ......................................................................TAAGAACTAGCTGCAGCAGTGTGTG........................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................CAGGATGGGGATGCTGAAAGC......................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGTGTGCTCATCCCGAAGTGACCTGCC......... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGTTGAATAAGAACTAGCTGCAGCAG................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTGTGTGTAGATCTGGGTGCACCAGG.......................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................GAATAAGAACTAGCTGCAGCAGTGTGT............................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CAGGATGGGGATGCTGAAAGCACAACCTA................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGTGTGCTCATCCCGAAGTGACCTGCCAGtt..... | 31 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTGTGTGTAGATCTGGGTGCACCAGGA......................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................TGTGTGTGCGTGCGTGCGTGCGTGCGTcc.................................................................................................................................... | 29 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGTAGATCTGGGTGCACCAGGATGGa..................................................................................... | 26 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................GTGCTCACTGTTGAATAAGAACTAGCTGCAG.................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................TGAATAAGAACTAGCTGCAGCAGTGT.............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TAAGAACTAGCTGCAGCAGTGTGTGT.......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGTTGAATAAGAACTAGCTGCAGCAGTG............................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTAGATCTGGGTGCACCAGGATGGG..................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................................CCCGAAGTGACCTGCCAGGCTCAGA | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................TGAATAAGAACTAGCTGCAGCAGTGTGT............................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTGCCCGTCTGCTGTTAGGTGCCAGCTATGCCCTGGCCCAGACAGTGACCGAAGTGTGCTCACTGTTGAATAAGAACTAGCTGCAGCAGTGTGTGTGCGTGCGTGCGTGCGTGCGTGCGTGTGCGTGTGCGTGTGTGTGTGTAGATCTGGGTGCACCAGGATGGGGATGCTGAAAGCACAACCTATCCTTGGTTTCCTAGCTCAATGGTGGCTCTGTGTGCTCATCCCGAAGTGACCTGCCAGGCTCAGA .............................................................(((((((....((......)).)))))))..((.((((..((..((((((..((((....))))..))))))..))..)))).))((((.....))))........................................................................................... .............................................................62....................................................................................................164.................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................GTGCGTGCGTGCGTGCGTGTGCGTGTGt......................................................................................................................... | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................CCGAAGTGTGCTCACTGTTGAATA.................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGCCCGTCTGCTGTTAGGTGCCAGCTA.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GCTCACTGTTGAATAAGAACTAGCTGCA..................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CGTGCGTGCGTGCGTGCGTGCGTGTatt................................................................................................................................ | 28 | att | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |