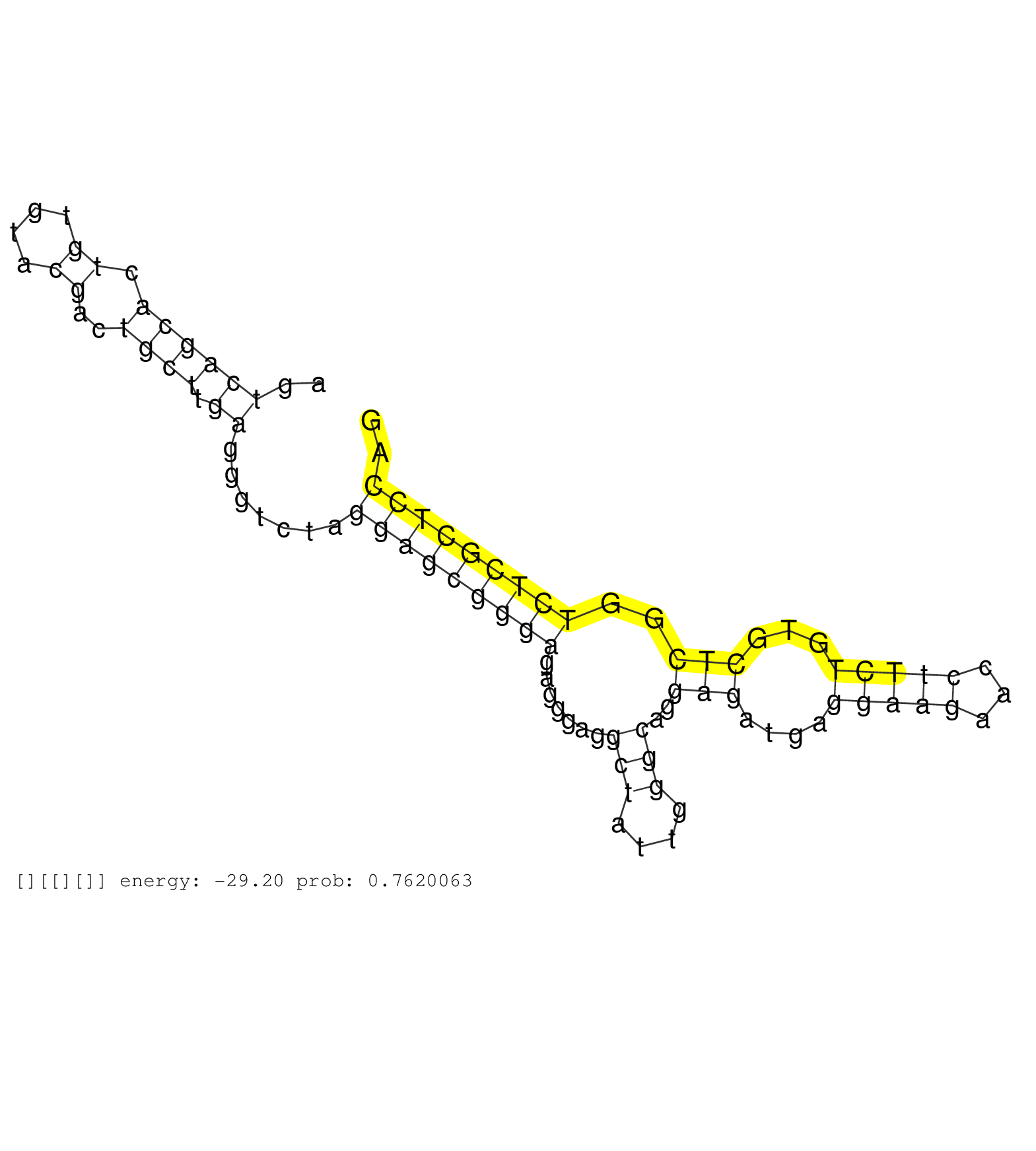
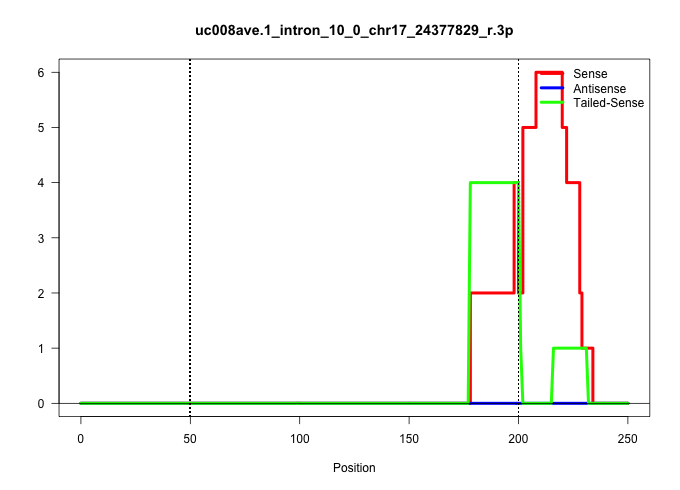
| Gene: Ccnf | ID: uc008ave.1_intron_10_0_chr17_24377829_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) PIWI.ip |
(14) TESTES |
| GCCACCGTCCGTGCTGCTTAGAGCAGAGCCCCGAGGCCAGGCTCCCTATTGGTCAGTTACCGAGCACTGGCTGCTTCTCAGCCTCATTTCTGCCGTGATCAGTCAGCACTGTGTACGACTGCTTGAGGGTCTAGGAGCGGGAGAGGGAGGCTATTGGGCAGGAGATGAGGAAGAACCTTCTGTGCTCGGTCTCGCTCCAGGACGAAGAGAAAAGGAAGCAGGCGCGTAGCCTTTTGGAAGAGTCTTCTCG ......................................................................................................((((((.((....))..)))).)).......(((((((((.......(((....)))..(((....(((((...)))))...)))..))))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TCTGTGCTCGGTCTCGCTCCAG.................................................. | 22 | 1 | 4.00 | 4.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................................................TCTGTGCTCGGTCTCGCTCCAGta................................................ | 24 | ta | 3.00 | 4.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTGTGCTCGGTCTCGCTCCAGt................................................. | 23 | t | 3.00 | 4.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................CGAAGAGAAAAGGAAGCAGGCGCGTA...................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCTGTGCTCGGTCTCGCT...................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTGTGCTCGGTCTCGCTCCAGa................................................. | 23 | a | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GAAAAGGAAGCAGGCGCGTAGCCTTT................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................AGGACGAAGAGAAAAGGAAGCA.............................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGACGAAGAGAAAAGGAAGCAGG............................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................................AGCAGGCGCGTAaatt.................. | 16 | aatt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CGAAGAGAAAAGGAAGCAGGCGCGTAG..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GCCACCGTCCGTGCTGCTTAGAGCAGAGCCCCGAGGCCAGGCTCCCTATTGGTCAGTTACCGAGCACTGGCTGCTTCTCAGCCTCATTTCTGCCGTGATCAGTCAGCACTGTGTACGACTGCTTGAGGGTCTAGGAGCGGGAGAGGGAGGCTATTGGGCAGGAGATGAGGAAGAACCTTCTGTGCTCGGTCTCGCTCCAGGACGAAGAGAAAAGGAAGCAGGCGCGTAGCCTTTTGGAAGAGTCTTCTCG ......................................................................................................((((((.((....))..)))).)).......(((((((((.......(((....)))..(((....(((((...)))))...)))..))))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................CTCGGTCTCGCTCtagt..................................................... | 17 | tagt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |