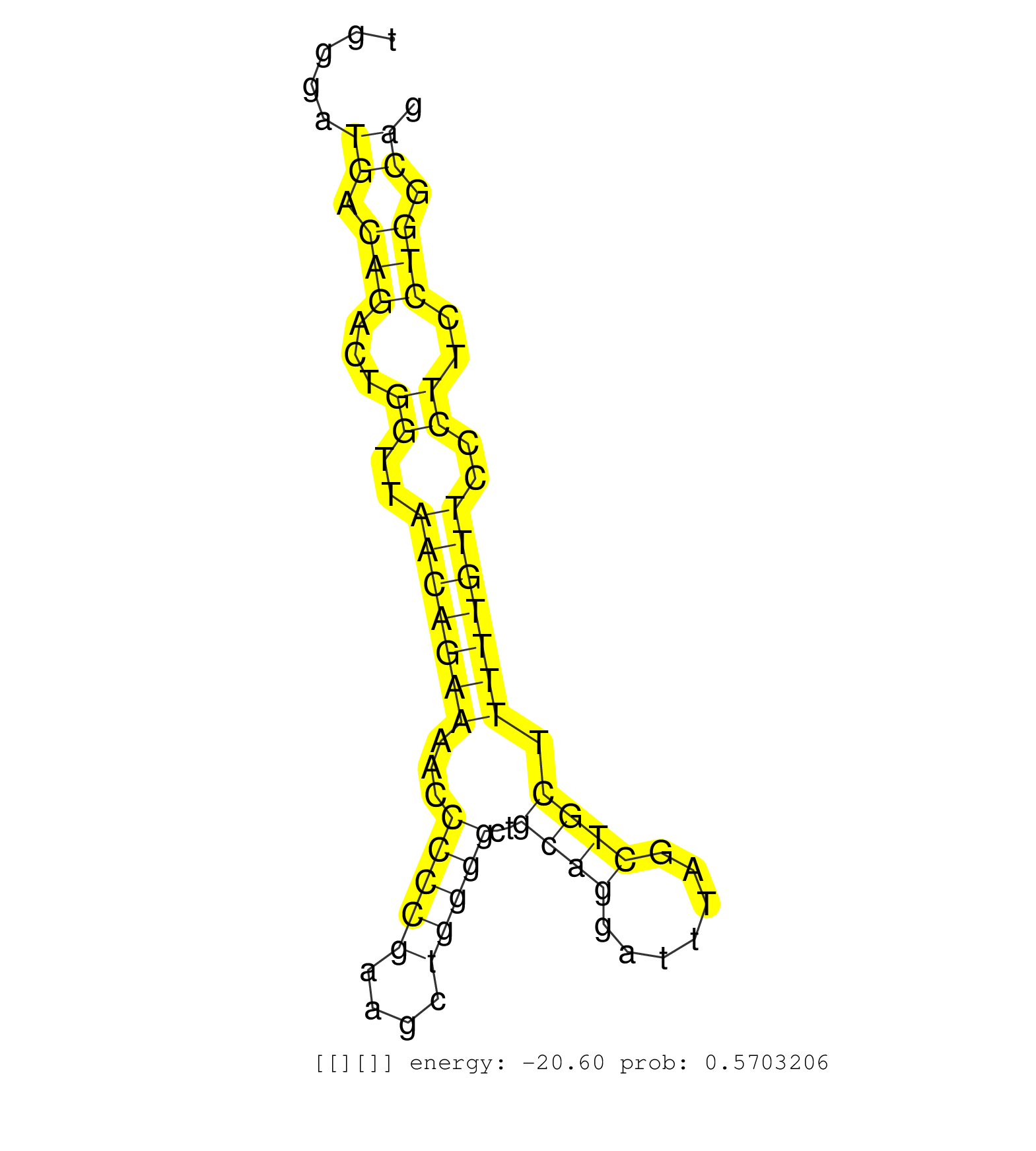

| Gene: Tbc1d24 | ID: uc008auq.1_intron_7_0_chr17_24323147_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(6) PIWI.ip |
(3) PIWI.mut |
(21) TESTES |
| CCATTATTTATCTTTCTAGGCCTTTTTGGAAAAATGAAAAAAACAAAACAAAACCCAAGTCCTTTGAAATGTGTACTTTCAATGCATTCACTAAAATGGTTTTCAATCCTACAGGGTGTGTGGGATGACAGACTGGTTAACAGAAAACCCCCGAAGCTGGGGCTGCAGGATTTAGCTGCTTTTTGTTCCCTTCCTGGCAGCCTAGGACCTGCCTTGTGGGCACACTGGGCACCATGGACCCCCCAGGGTA .............................................................................................................................((.(((...((..(((((((...(((((....)))))..((((.......)))).)))))))..))..))).))................................................... ........................................................................................................................121............................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................TGACAGACTGGTTAACAGAAAACCCCC.................................................................................................. | 27 | 1 | 5.00 | 5.00 | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................TGGGATGACAGACTGGTTAACAGAAAAC...................................................................................................... | 28 | 1 | 5.00 | 5.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGGGATGACAGACTGGTTAACAGAAAA....................................................................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAGGACCTGCCTTGTGGGCACACTGGGC.................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................GACAGACTGGTTAACAGAAAACCCCC.................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGTGGGATGACAGACTGGTTAACAGAA......................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TAGCTGCTTTTTGTTCCCTTCCTGGC.................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGACAGACTGGTTAACAGAAAACCCCCGAAatt............................................................................................ | 33 | att | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGTGGGATGACAGACTGGTTAACAGAAA........................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CGAAGCTGGGGCTGCAGGATTTAGC.......................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................AAAACCCCCGAAGCTGGGGCTGCAGGA................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TTTAGCTGCTTTTTGTTCCCTTCCTGG..................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................TTTAGCTGCTTTTTGTTCCCTTCCTGGC.................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................AATGGTTTTCAATCCTACAGGGTGTGTG................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGGGATGACAGACTGGTTAACAGAAAACC..................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGACAGACTGGTTAACAGAAAACCCCCGA................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................TAAAATGGTTTTCAATCCTACAGGGTGTGT................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGGGATGACAGACTGGTTAACAGAAAt....................................................................................................... | 27 | t | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GGATTTAGCTGCTTTTTGTTCCCTTC......................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TAACAGAAAACCCCCGAAGCTGGGGCTGCAG.................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............TTTCTAGGCCTTTTTGGAAAAATGAAA................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................TAACAGAAAACCCCCGAAGCTGGGGCTGCAGG................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TACTTTCAATGCATTCACTAAAATGGTTTT................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGACAGACTGGTTAACAGAAAACCCCCt................................................................................................. | 28 | t | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTTCAATCCTACAGatgc.................................................................................................................................... | 18 | atgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................TGGGATGACAGACTGGTTAACAGAAA........................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GTGTGTGGGATGACAGACTGGTTAACA............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGACAGACTGGTTAACAGAAAACCCC................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGGCACACTGGGCACCATGGACCCCCC...... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCCCGAAGCTGGGGCTGCAGGATTTA............................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................GTGGGATGACAGACTGGTTAACAGAAA........................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................TGGGATGACAGACTGGTTAACAGAAAACac.................................................................................................... | 30 | ac | 1.00 | 5.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCATTATTTATCTTTCTAGGCCTTTTTGGAAAAATGAAAAAAACAAAACAAAACCCAAGTCCTTTGAAATGTGTACTTTCAATGCATTCACTAAAATGGTTTTCAATCCTACAGGGTGTGTGGGATGACAGACTGGTTAACAGAAAACCCCCGAAGCTGGGGCTGCAGGATTTAGCTGCTTTTTGTTCCCTTCCTGGCAGCCTAGGACCTGCCTTGTGGGCACACTGGGCACCATGGACCCCCCAGGGTA .............................................................................................................................((.(((...((..(((((((...(((((....)))))..((((.......)))).)))))))..))..))).))................................................... ........................................................................................................................121............................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................ACTGGTTAACAGAAAAaag....................................................................................................... | 19 | aag | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |