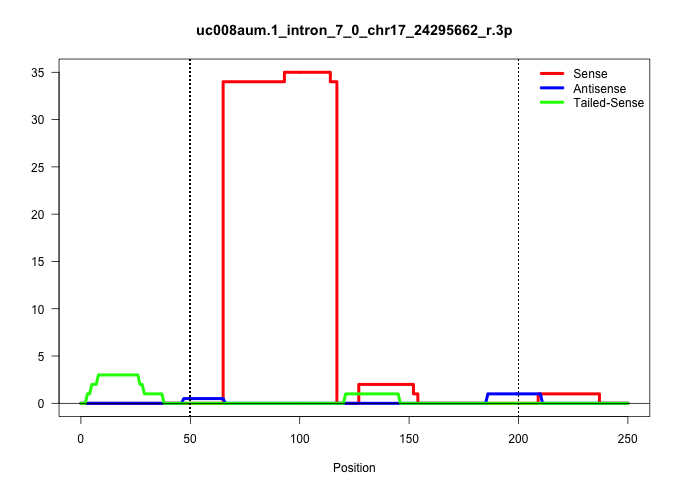
| Gene: Amdhd2 | ID: uc008aum.1_intron_7_0_chr17_24295662_r.3p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(1) PIWI.ip |
(1) TDRD1.ip |
(11) TESTES |
| CCTTGTGGTAGGTCATCAAGTAGTCCTGGTTGTCAGGTCTTCCTGCACCTGAGCACTGGGGGAGGAGGGCCTTCATCTTCAGCTCCTGTGGGGAAGGCCAGTCGGGCTGGGCTGGGTCAGATGAGGATCTGACCTTGTTGGGCTGTGGCTTCTGGGTGCTGGGCTGTGGGGACAGGGCTTTAACAGCTGGTTCCCCCCAGGTCCTTCCTCAGATCCCTGTGAAGAGTGGAGGTCCCCATGGGGCAGGGGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................AGGGCCTTCATCTTCAGCTCCTGTGGGGAAGGCCAGTCGGGCTGGGCTGGGT..................................................................................................................................... | 52 | 1 | 34.00 | 34.00 | 34.00 | - | - | - | - | - | - | - | - | - | - |
| ........TAGGTCATCAAGTAGTCCTGGTTGTCAGGg.................................................................................................................................................................................................................... | 30 | g | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................AAGGCCAGTCGGGCTGGGCTG........................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...TGTGGTAGGTCATCAAGTAGTCCc............................................................................................................................................................................................................................... | 24 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................CAGATCCCTGTGAAGAGTGGAGGTCCCC............. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................TCTGACCTTGTTGGGCTGTGGCTTC.................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................TCTGACCTTGTTGGGCTGTGGCTTCTG................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGAGGATCTGACCTTGTTGGGCTGa........................................................................................................ | 25 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....TGGTAGGTCATCAAGTAGTCCcgg............................................................................................................................................................................................................................. | 24 | cgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| CCTTGTGGTAGGTCATCAAGTAGTCCTGGTTGTCAGGTCTTCCTGCACCTGAGCACTGGGGGAGGAGGGCCTTCATCTTCAGCTCCTGTGGGGAAGGCCAGTCGGGCTGGGCTGGGTCAGATGAGGATCTGACCTTGTTGGGCTGTGGCTTCTGGGTGCTGGGCTGTGGGGACAGGGCTTTAACAGCTGGTTCCCCCCAGGTCCTTCCTCAGATCCCTGTGAAGAGTGGAGGTCCCCATGGGGCAGGGGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................CTGGTTCCCCCCAGGTCCTTCCTCA....................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................CCTGAGCACTGGGGGAGGA........................................................................................................................................................................................ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 |