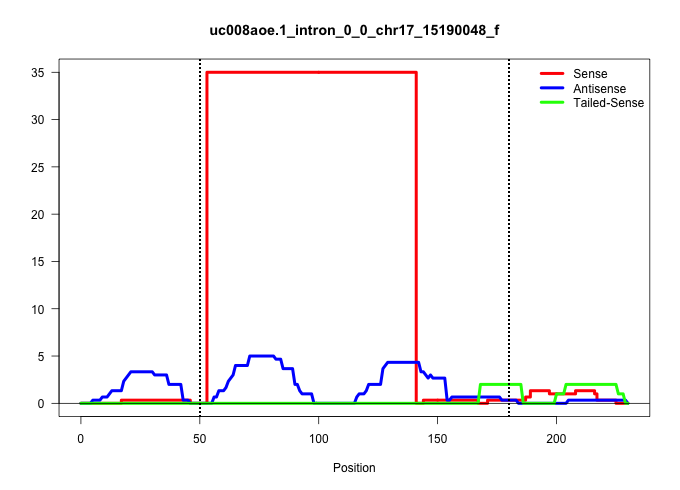
| Gene: 9030025P20Rik | ID: uc008aoe.1_intron_0_0_chr17_15190048_f | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(2) PIWI.mut |
(28) TESTES |
| AGGCTGTACTGAACAAGGGCTCCCCACTACAGTTCCAGCTGCAGGATGGGGTGAGTGCCCTCTTTGAGACTTGCATTTCCCATCAGTTTATAAACAAGCCAAGAACTGCCAGGATACCCTGCCATTCACTCTTCATCTGCCCAGAACACTGACAGAATGTTTCTCTGTCAATTCTAAAAGGAATTCTTGTGGGATTTCCTGAACTATCAGGAGGGTCCTCGTATCCGGGA ..................................................(((((((..(((...)))....(((..(((...((((((.............))))))...))).....))))))))))......(((....((((...(((((((.......))))))).))))....)))................................................ ..................................................51......................................................................................................................................187......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................AGTGCCCTCTTTGAGACTTGCATTTCCCATCAGTTTATAAACAAGCCAAGAA............................................................................................................................. | 52 | 1 | 36.00 | 36.00 | 36.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CAATTCTAAAAGGAgcga............................................ | 18 | gcga | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CCCCACTACAGTTCCAGCTGCA........................................................................................................................................................................................... | 22 | 3 | 1.67 | 1.67 | - | - | - | - | - | - | - | - | - | - | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TATCAGGAGGGTCCTCGTATCCGGt. | 25 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGCCCTCTTTGgaa................................................................................................................................................................. | 18 | gaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAACTATCAGGAGGGTCCTCGTATCt.... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGGGATTTCCTGAACTATCAGGAGGGTC............. | 28 | 3 | 0.67 | 0.67 | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AGGAGGGTCCTCGTATC..... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TGTGGGATTTCCTGAACTATCAGGAGGGT.............. | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTCTAAAAGGAATTCTTGTGGGATTT................................. | 26 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AACACTGACAGAATGTTTCTCTG............................................................... | 23 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - |
| .................GGCTCCCCACTACAGTTCCAGCTGCAGGA........................................................................................................................................................................................ | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| AGGCTGTACTGAACAAGGGCTCCCCACTACAGTTCCAGCTGCAGGATGGGGTGAGTGCCCTCTTTGAGACTTGCATTTCCCATCAGTTTATAAACAAGCCAAGAACTGCCAGGATACCCTGCCATTCACTCTTCATCTGCCCAGAACACTGACAGAATGTTTCTCTGTCAATTCTAAAAGGAATTCTTGTGGGATTTCCTGAACTATCAGGAGGGTCCTCGTATCCGGGA ..................................................(((((((..(((...)))....(((..(((...((((((.............))))))...))).....))))))))))......(((....((((...(((((((.......))))))).))))....)))................................................ ..................................................51......................................................................................................................................187......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................GCCATTCACTCTTCATCTGCCCAGAACA.................................................................................. | 28 | 3 | 3.00 | 3.00 | - | - | - | - | 0.33 | 2.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CTCCCCACTACAGTTCCAGCTGCA........................................................................................................................................................................................... | 24 | 3 | 3.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | 0.33 | 0.33 | - | - | - | - | 1.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............GGGCTCCCCACTACAGTTCCAGCTa.............................................................................................................................................................................................. | 25 | a | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GCTCCCCACTACAGTTCCAGCTGCA........................................................................................................................................................................................... | 25 | 3 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 0.33 | 1.00 | - | - | - | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| ...............................................................................................................................ACTCTTCATCTGCCCAGAACACTGACA............................................................................ | 27 | 3 | 1.67 | 1.67 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ....................TCCCCACTACAGTTCCAGCTGCA........................................................................................................................................................................................... | 23 | 3 | 1.67 | 1.67 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CCCTGCCATTCACTCTTCATCTGCCCA....................................................................................... | 27 | 3 | 1.00 | 1.00 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| .......................................................................TGCATTTCCCATCAGTTTATAAACAAG.................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CTTGCATTTCCCATCAGTTTAactc............................................................................................................................................ | 25 | actc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CCCACTACAGTTCCAGCTGCAGGA........................................................................................................................................................................................ | 24 | 3 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CATTCACTCTTCATCTGCCCAGAaaa..................................................................................... | 26 | aaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CCTCTTTGAGACTTGCATTTCCCATCA................................................................................................................................................. | 27 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| ............ACAAGGGCTCCCCACTACAGTTCCA................................................................................................................................................................................................. | 25 | 3 | 0.67 | 0.67 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TTGAGACTTGCATTTCCCATCAGTTTA............................................................................................................................................ | 27 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GGCTCCCCACTACAGTTCCAGCTGCA........................................................................................................................................................................................... | 26 | 3 | 0.67 | 0.67 | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CCCCACTACAGTTCCAGCTGCAGGA........................................................................................................................................................................................ | 25 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........ACAAGGGCTCCCCACTACAGTTCCAa................................................................................................................................................................................................. | 26 | a | 0.67 | 0.00 | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GAGACTTGCATTTCCCATCAGTTTATA.......................................................................................................................................... | 27 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - |
| ..............................................................TTTGAGACTTGCATTTCCCATCAGTTTA............................................................................................................................................ | 28 | 3 | 0.67 | 0.67 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GCCCTCTTTGAGACTTGCATTTCCCATCA................................................................................................................................................. | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GAGACTTGCATTTCCCATCAGTTTATAA......................................................................................................................................... | 28 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....GTACTGAACAAGGGCTCCCCACTACA....................................................................................................................................................................................................... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| .....................................................................................................................CCTGCCATTCACTCTTCATCTGCCCA....................................................................................... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGAACAAGGGCTCCCCACTACAGTTCCA................................................................................................................................................................................................. | 28 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................ATCAGGAGGGTCCTCGTATCCGGGA | 25 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................ATGTTTCTCTGTCAATTCTAAAAGGAAT.............................................. | 28 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GAACAAGGGCTCCCCACTACAGTTCCA................................................................................................................................................................................................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTTCATCTGCCCAGAACACTGACA............................................................................ | 25 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| .............................................................CTTTGAGACTTGCATTTCCCATCAGTTTA............................................................................................................................................ | 29 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TGAGACTTGCATTTCCCATCAGTTTA............................................................................................................................................ | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............CAAGGGCTCCCCACTACAGTTCCA................................................................................................................................................................................................. | 24 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGAGACTTGCATTTCCCATCAGTTTAccga............................................................................................................................................ | 30 | ccga | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................CCATTCACTCTTCATCTGCCCAGA..................................................................................... | 24 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................CCATTCACTCTTCATCTGCCCAGAA.................................................................................... | 25 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CTCTTCATCTGCCCAGAACACTGACA............................................................................ | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| ........................................................GCCCTCTTTGAGACTTGCATTTCCCA.................................................................................................................................................... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................ACTGACAGAATGTTTCTCTGTCAATTCTAA..................................................... | 30 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CCCCACTACAGTTCCAGCTGCA........................................................................................................................................................................................... | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAACACTGACAGAAT........................................................................ | 17 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |